Rice is a staple food for more than half of the human population. It has been estimated that by 2030, rice production must increase by 40% to meet the growing demand (Khush, 2005). In addition, with the improvement of people’ s living standards, the demand for elite rice with better eating and cooking quality (ECQ) is increasing. ECQ is determined by several factors, including amylose content (AC), gel consistency (GC), gelatinization temperature (GT) and viscosity, where AC is the predominant factor (Juliano, 1998).
Amylose is synthesized by GRANULE-BOUND STARCH SYNTHASE I (GBSSI), which is encoded by the Waxy (Wx) gene (LOC_Os06g04200) (Wang et al, 1995). Allelic variations in Wx contribute to differences in AC. The major alleles Wxa and Wxb, from indica and japonica varieties respectively, differed by a G/T substitution that leads to differential splicing between Wxa andWxb(Isshiki et al, 1998). The expression of Wxa is around 10-fold higher than that of Wxb, resulting in indica varieties having higher AC than japonicavarieties (Isshiki et al, 1998). Wxop, Wxmp, Wxmq and Wxhp are partially functional alleles of Wx. The AC in rice varieties with Wxop, Wxmp and Wxhp genotypes is approximately 7%-10%, leading to reduced transparency of milled rice and softer texture in cooked rice (Zhu et al, 2015). These cultivars are therefore known as soft rice. In addition, cultivars with loss-of-function alleles of Wx are called waxy rice or glutinous rice, and are very sticky when cooked.
In the past decade, efforts to improve ECQ by reducing AC have focused on Wx manipulation (Liu et al, 2003, 2005). For example, transfer of the Wxb allele into the indica cultivar significantly improved its ECQ (Liu et al, 2006). In addition, theWxmq allele has been widely used in japonica breeding in Jiangsu Province, China and several resulting cultivars are very popular in this region (Yao et al, 2017). However, to transfer the Wxb and Wxmp genotypes, breeders have to use traditional breeding methods involving multiple rounds of backcrossing, which is time-consuming and laborious (Yao et al, 2017). Recently, the CRISPR/Cas9 (clustered regulatory interspaced short palindromic repeat/CRISPR-associated protein 9) system has proven to be a powerful, rapid and cost-effective approach for rice improvement and breeding. Agronomic traits such as seed shape, architecture, heading date, herbicide resistance and fragrance have been modified successfully using CRISPR/Cas9 in rice (Li et al, 2016; Sun et al, 2016; Li et al, 2017). The Wx gene has also been edited to produce glutinous rice without loss of yield (Ma et al, 2015; Zhang et al, 2018; Fei et al, 2019).
In practice, glutinous rice is a specialty product, with a small market compared to common rice. Soft rice with 7%-10% of AC is more popular in South China than in North China, where people feel the soft rice is too sticky. In addition, the dark and cloudy endosperm of the soft rice cultivar is considered undesirable in appearance. Therefore, we aimed to create soft but less sticky rice by controlling AC to within 10%-12% (a value between that of japonica and soft rice). However, there were few native Wx alleles available for use.
Given this limitation, we set out to generate new Wx alleles via the CRISPR/Cas9-mediated gene editing. The Wxmp and Wxmqalleles encode proteins that share an amino acid substitution (R158H) responsible for soft rice, but have a different single nucleotide polymorphism (SNP) in the exon 5 of the Wx gene (Sato et al, 2002; Yang et al, 2013; Zhou et al, 2020). We hypothesized that this amino acid site might be critical for Wx function and chose the Wxmp-containing region as the target for CRISPR/Cas9-mediated base editing. We aimed to create an amino acid substitution mutation close to R158H and thereby generate novel Wx alleles that might slightly reduce AC without affecting grain appearance.
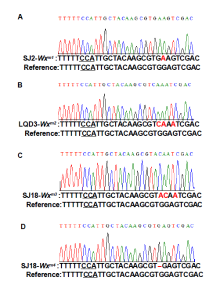
We first designed single guide RNAs (sgRNAs) to target the Wxmp-containing region (Fig. 1-A). There are several guanine (G) bases close to the putative protospacer-adjacent motif (PAM) site (NGG) of Wxmp, and we thus used cytidine base editors (CBEs), which have high editing efficiency in rice (Wang et al, 2019). The sgRNA was ligated into a base editing vector Anc689BE4max. Suijing 18 (SJ18), Songjing 2 (SJ2), and Longqingdao 3 (LQD3), three elite rice varieties widely grown in Heilongjiang Province, China, were used for transformation. A total of 18 independent transgenic lines were generated at the T0 generation (Supplemental Fig. 1).
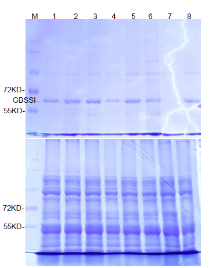
Mutations in the Wx target region were identified through sequencing the flanking region and decoded by the Degenerate Sequence Decoding method (Liu et al, 2015). Fourteen seedlings containing G-to-A and G-to-C substitutions were chosen for further analysis. In the T1 generation, we first identified the transgene-free plants through PCR assays with hygromycin- and CAS9-specific primers (Supplemental Table 1). The transgene- free plants were further examined for homozygous substitution types by sequencing. Finally, the transgene-free and homozygous substitution lines were identified (Supplemental Fig. 2). Most of the mutations were G-to-A substitutions before PAM (Fig. 1-B and Supplemental Fig. 2). Diverse mutation types were generated due to the number and position of G substitutions in the target region. The SJ2, LQD3 and SJ18 backgrounds gave rise to one substitution line each: Wxm1 (G159E), Wxm2(G159Q and V160I) and Wxm3(G159T and V160I), respectively. In addition, we identified a loss-of-function allele (Wxm4) in SJ18 caused by a 1-bp deletion, similar to the waxy mutation (Fig. 1-B and Supplemental Fig. 2). To examine whether the protein levels were changed in these Wx-edited lines, we extracted GBSSI protein and examined it in SDS-PAGE gels. Compared with their parents, the GBSSI protein levels were decreased in Wxm2 and Wxm3, and diminished in Wxm4, suggesting that Wx protein expression levels were changed in theseWx-edited lines (Supplemental Fig. 3).
| Supplemental Table 1. Primers used in this study. |
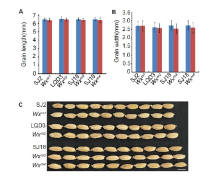
During growth in greenhouse, we monitored agronomic traits, including growth speed, growth vigor and heading date, and observed that the edited lines Wxm1 to Wxm4 were indistinguishable from their parents. These results indicated that the editing of Wx did not affect the rice growth and development. After gathering the ripe seeds, the rice grains were husked. There were no observable differences between the parent and edited lines Wxm1 to Wxm3, including the traits of color and lustre, transparency and chalkiness, which indicated that these Wx-edited linesWxm1 toWxm3 were not affected in appearance quality (Fig. 1-C and Supplemental Fig. 4). By contrast, the edited Wxline Wxm4appeared milky white and opaque (Fig. 1-C), similar to glutinous rice (Zhang et al, 2018; Fei et al, 2019).
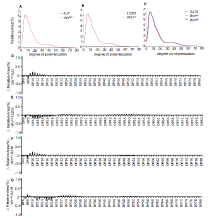
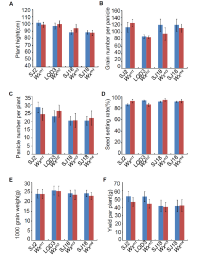
Notably, the apparent ACs in the edited lines (Wxm1 toWxm3) were slightly but significantly lower (4.80%-13.41%) than those in the parents (Fig. 1-D), which is consistent with the decreased Wx protein levels in these lines. This result demonstrated that amino acid substitutions close to the one in Wxmp indeed led to impaired function of the Wx gene product, decreasing the amount of amylose in the seed. By contrast, the loss-of-function Wxm4line showed a much lower AC (Fig. 1-D), similar to what was previously observed (Zhang et al, 2018). In addition, the amylopectin chain length distributions in the edited lines were similar to those in the parents, indicating that the amylopectin structure was unchanged (Supplemental Fig. 5). Further, there were no observable differences in yield-related traits, including plant height, grain number per panicle, panicle number per plant, seed-setting rate, 1000-grain weight and yield per plant, between the edited lines and parents (Supplemental Fig. 6). Thus, we concluded that the yields of these Wx gene-edited lines were not affected, which is similar to the results in a previous study (Zhang et al, 2018). In future work, we plan to harvest more edited seeds to further evaluate their ECQ, and to grow these edited lines in paddy fields at larger scale to further evaluate their performance and value for practical rice breeding and production.
In summary, we used the CRISPR/Cas9-mediated base editing system to develop a simple and highly efficient approach for mild reduction of rice AC, by which the rice ECQ might be improved. To our knowledge, this is the first report about moderately reducing rice AC via base editing of the Wx gene. Our approach of slightly decreasing Wx function is different from previous studies that generated wx lines via base insertion or deletion of Wx genes, and thereby fully disrupting Wx function (Ma et al, 2015; Zhang et al, 2018; Fei et al, 2019). Importantly, the appearance, quality and yield-related traits of edited lines were not impaired. All these transgene-free and homozygous lines were generated in less than one year, which saves a significant amount of time and labor compared to traditional breeding method. This study also indicated that it is feasible to create beneficial mutations by editing amino acids close to the critical domain of the product of a target gene.
This research was supported by the National Transgenic Science and Technology Program (Grant No. 2018ZX0800102B), National Key Research and Development Program of China (Grant No. 2017YFD0100501), National Natural Science Foundation of China (Grant No. 31801327), Key Research and Development Program of Jiangsu Province (Grant No. BE2017345-2) and Natural Science Foundation of Heilongjiang (Grant No. C2018065). We thank Prof. Zhu Jiankang of Shanghai Center for Plant Stress Biology, Chinese Academy of Sciences for providing the base editing vector.
The following materials are available in the online version of this article at http://www.sciencedirect.com/science/journal/16726308; http://www.ricescience.org.
Supplemental File 1. Materials and methods.
Supplemental Fig. 1. Identification of transgenic plants in T0 generation through PCR assays with hygromycin gene- specific primers.
Supplemental Fig. 2. Identification of edited Wx mutants.
Supplemental Fig. 3. Detection of GBSSI in parents and edited lines.
Supplemental Fig. 4. Grain morphology in parents and edited lines.
Supplemental Fig. 5. Amylopectin structure analysis in parents and edited lines.
Supplemental Fig. 6. Agronomic trait analysis in parents and edited lines.
Supplemental Table 1. Primers used in this study.
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|