#These authors contributed equally to this work
Aluminum (Al) toxicity in acid soils is a significant limitation to crop production worldwide, as 13% of the world’s rice is produced in acid soil with high Al content. Rice is likely the most Al-resistant cereal and also the cereal, where Al resistance is the most genetically complex with external detoxification and internal tolerance. Many Al-resistance genes in rice have been cloned, including Al resistance transcription factor 1 ( ART1) and other transcription factors, organic acid transporter genes, and metal ion transporter gene. This review summarized the recent characterized genes affecting Al tolerance in rice and the interrelationships between Al and other plant nutrients.
Aluminum (Al) is the third most abundant element in the earth crust, after oxygen and silicon, and the most plentiful metallic element in soil. About 40% of arable soil in the world has high acidity (Ma, 2000; Kochian et al, 2004). In acid soil, where the soil pH is commonly below 5.0, many soluble ionic Al (mainly Al3+) is released (Ryan et al, 2001; Kochian et al, 2005; Ma, 2007). Al3+ is a very reactive element and binds to multiple subcellular organelles including the cell wall, plasma membrane, cytoskeleton and nucleus, and then affects their functions (Ma et al, 2014; Kochian et al, 2015). Therefore, Al toxicity is one of the most important factors limiting plant growth and crop yield in acid soil, and the harm becomes more and more obvious with the increase of acid rain and the use of chemical fertilizer (Kochian et al, 2004; Ma, 2007; Huang et al, 2012; Roy and Bhadra, 2014).
Rice (Oryza sativa) is one of the most important food crops in the world. About 13% of the world’ s rice is produced in acid soil. Compared with other crops, rice has relatively stronger Al toxic resistance (Famoso et al, 2010, 2011), and is also the most complex cereal crop with Al resistance genes (Wu et al, 2000; Ma et al, 2002; Nguyen et al, 2003). Nevertheless, as for other crops, Al toxicity limits rice growth and mineral element absorption and subsequently reduces grain yield (Ma et al, 2002; Huang et al, 2009).
Al tolerance in rice is controlled by multiple genes and many QTLs associated with Al tolerance have been detected through linkage mapping and genome- wide association studies (Wu et al, 2000; Ma et al, 2002; Nguyen et al, 2003; Famoso et al, 2011; Meng et al, 2017; Tao et al, 2018; Zhao et al, 2018). A number of genes have been well characterized using reverse genetics approach, which greatly enhances our understanding of the molecular mechanisms of rice response to Al toxicity. The interrelationships between Al and other plant nutrients have gained attention and provide another angle to gain insights of the complexity of Al tolerance. In this review, we briefly reviewed the advances by summarizing newly discovered and characterized genes.
Plants can develop several Al resistance mechanisms to overcome the stress caused by Al toxicity. These mechanisms can be categorized as external detoxification and internal tolerance (Ma et al, 2014; Kochian et al, 2015; Zhang X et al, 2019). The well-studied plant strategy of external detoxification is where roots can exclude Al by secreting organic acids (Ma et al, 2001, 2007, 2014; Kochian et al, 2004, 2015; Yang et al, 2008; Zhu et al, 2012). In contrast, the internal tolerance strategies allow the plant to tolerate Al accumulation either in the root cell wall by binding Al to pectin and hemicellulose (Schmohl et al, 2000; Zhu et al, 2012; Yang et al, 2013; Xia et al, 2013; Ma et al, 2014), or in root symplasm via Al uptake and chelation/sequestration (Xia et al, 2010; Huang et al, 2012; Ma et al, 2014; Kochian et al, 2015). Fig. 1 outlines the current understanding of molecular mechanisms of Al tolerance in rice.
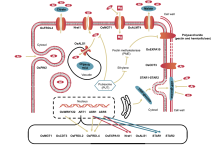 | Fig. 1. Mechanisms for aluminum (Al) tolerance in rice. OsWRKY22, WRKY transcription factor (Li G Z et al, 2018); ART1, Al resistance transcription factor 1 (Yamaji et al, 2009); ASR1, ABA-stress and ripening 1 (Arenhart et al, 2016); ASR5, ABA-stress and ripening 5 (Arenhart et al, 2012, 2013); OsMGT1, Magnesium transporter (Chen Z C et al, 2012); OsCDT3, A small peptide with rich cysteine (Xia et al, 2013); OsFRDL2/OsFRDL4, Citrate transporter (Yokosho et al, 2011); OsEXPA10, An expansin gene (Che et al, 2016); Nrat1, Nramp aluminum transporter 1 (Xia et al, 2010); OsALS1, Tonoplast-localized Al transporter (Yamaji et al, 2009); STAR1/STAR2, UDP-glucose transporter (Huang et al, 2009, 2012); OsPIN2, An auxin efflux transporter (Wu et al, 2014); OsALMT4, Malate transporter (Liu J et al, 2017). |
The reprogramming of gene expression through transcriptional regulation, mediated by transcription factors, is a finely orchestrated and tightly regulated process, and is one of the hallmarks of plant response to stress (Vaahtera and Brosché , 2011). To translate a stress exposure into appropriate changes in gene expression, suitable signalling pathways need to be activated, ultimately ending up with a transcription factor at the promoter of the gene to be transcribed (Vaahtera and Brosché , 2011).
The first transcript factor identified for Al tolerance in rice is the Al resistance transcription factor 1, ART1 (Yamaji et al, 2009; Arbelaez et al, 2017). ART1 is a C2H2 zinc-finger protein, which is a homolog of STOP1 in Arabidopsis thaliana (Tsutsui et al, 2011). STOP1 regulates the expression of AtMATE1 and AtALMT1 (Liu et al, 2009; Sawaki et al, 2009). ART1 binds to the core cis-acting element [GGN(T/g/a/C) V(C/A/g)S(C/G)], which presents in the promoter of ART1 downstream genes (Tsutsui et al, 2011), and regulates at least 31 genes (Yamaji et al, 2009), including STAR1, STAR2, OsFRDL4, OsCDT3, OsMGT1, Nrat1, OsALS1, OsFRDL2andOsEXPA10 (Table 1).
| Table 1 Lists of aluminum (Al) tolerance genes identified in rice. |
Functional analysis of some ART1 downstream genes showed that they are involved in the detoxification of Al both externally and internally (Fig. 1). STAR1 and STAR2 encode an ATP-binding domain and a membrane-binding domain, respectively, of a bacterial type ABC transporter (Huang et al, 2009; Arenhart et al, 2014). The STAR1-STAR2 complex localizes at the vesicles and transports UDP-glucose, which may be involved in cell wall modification, resulting in decreased Al accumulation in the cell wall (Huang et al, 2012). OsFRDL4, OsCDT3, OsMGT1 and Nrat1 all encode plasma-membrane-localized proteins. OsFRDL4is responsible for the secretion of citrate in response to Al toxicity (Yokosho et al, 2011). OsCDT3, encoding a small cysteine-rich peptide, shows binding activity with Al, thereby preventing Al entering into the root cells (Xia et al, 2013). OsMGT1 functions as a magnesium (Mg) transporter (Chen Z C et al, 2012; Zhang L D et al, 2019), and up-regulation of OsMGT1 can alleviate internal Al toxicity by enhancing Mg uptake (Chen Z C et al, 2012). Nrat1, a member of Nramp family, transports trivalent Al (Xia et al, 2010), which is required for sequestration of Al into the vacuoles for final detoxification. Vacuolar sequestration of Al is mediated by OsALS1, a half-size ABC transporter localized at the tonoplast (Huang et al, 2012). Among these genes examined, OsFRDL4 shows a good correlation between the expression and Al tolerance (Yokosho et al, 2011). OsFRDL2, encoding a MATE citrate transporter, and OsEXPA10, encoding an expansin for cell elongation in the root tips, are specifically induced by Al, but have only minor effects on Al tolerance (Che et al, 2016; Yokosho et al, 2016a).
In the rice genome, there are five close homologs of ART1 (Yamaji et al, 2009). Although ART2 is regulated by ART1, it regulates different genes related to Al tolerance, which indicates that ART2 is a supplementary pathway leading to Al tolerance in rice (Che et al, 2018).
The WRKY transcription factors, one of the largest families of transcription factors in plants, play crucial roles in the regulation of biotic stress, senescence and various developmental processes (Rushton et al, 2010; Jiang et al, 2017). The expression of OsFRDL4 is regulated by OsWRKY22 (Li G Z et al, 2018). In response to Al, OsWRKY22 promotes OsFRDL4 expression by binding to W-box cis-element within the promoter of OsFRDL4, and results in increased citrate secretion and Al tolerance. OsWRKY22 and ART1 jointly regulate Al-induced expression of OsFRDL4 and citrate secretion (Li G Z et al, 2018).
ASR (abscisic scid, stress and ripening) proteins have been identified exclusively in plants and have roles in response to abiotic (Arenhart et al, 2013; Hu et al, 2013; Joo et al, 2013) and biotic stresses (Liu et al, 2013). Acting as chaperones (Konrad and Bar-Zvi, 2008) and transcription factors (Arenhart et al, 2014; Ricardi et al, 2014), these proteins drive plant responses to environmental cues. The transcription factor ASR5 has been reported to be involved in Al tolerance in rice (Arenhart et al, 2012, 2013). ASR5 is localized in both nucleus and cytoplasm, and regulates the expression of different genes that collectively protect rice cells from Al-induced stress responses. Furthermore, ASR5 protein can bind STAR1 promoter and regulate STAR1, Nrat1 and OsFRDL4 (Arenhart et al, 2013, 2014). ASR1 has the same subcellular localization as ASR5; binds to ASR5 cis-regulatory elements; regulates ASR5- regulated genes in a non-preferential manner and might replace ASR5 under certain conditions (Arenhart et al, 2016). These results indicate that ASR1 and ASR5 act in concert and complementarily regulate the expression of genes in response to Al.
It is well-known that root apex is the critical region of Al toxicity. Many plants secrete organic acid from the root tip in response to Al stress, which prevents Al3+ from entering root tip cells (Fan et al, 2014; Ma et al, 2014; Kochian et al, 2015; Awasthi et al, 2019). OsFRDL4, belonging to the MATE family, is an Al-induced citrate transporter (Yokosho et al, 2011). It is localized at the plasma membrane of rice root cells and responsible for releasing citrate from the roots in response to Al toxicity. Recently, it was shown that a 1.2 kb transposon insertion at the promoter region is required for enhancing the expression of OsFRDL4, and this insertion is present in most japonica cultivars (Yokosho et al, 2016b). ART1 specifically regulates the expression of OsFRDL4 in rice root tip, and promotes the secretion of citric acid to chelate external Al ions and thus reduces Al toxicity (Yokosho et al, 2016b).
OsFRDL2 protein is localized at unidentified vesicles in the cytosol, and exhibits an efflux transport activity for citrate. OsFRDL2 is mainly expressed in the roots and its expression is rapidly up-regulated by Al. Furthermore, the expression of OsFRDL2 is also regulated by ART1. Knockout of OsFRDL2 decreases Al-induced secretion of citrate from the roots without affecting the internal citrate concentration. Al-induced inhibition of root elongation is similar between the OsFRDL2 knockout line and its wild type. Collectively, these results indicate that although OsFRDL2 is involved in the Al-induced secretion of citrate, its contribution to high Al tolerance is relatively small in rice (Yokosho et al, 2016a).
Aluminum-activated malate transporter (ALMT) family genes encode anion channels in plants (Delhaize et al, 2004, 2007; Barbier-Brygoo et al, 2011; Dreyer et al, 2012; de Angeli et al, 2013). The family is named after the first member identified, TaALMT1, which controls the major mechanism for Al resistance in wheat (Sasaki et al, 2004). TaALMT1 is expressed in the root apices of Al-resistant wheat germplasm, and in acid soil, the high concentration of soluble Al3+ activates the channel protein to release malate anions into the apoplast. Malate anions then bind with the toxic Al3+ to protect root cells and maintain root growth (Pineros et al, 2008; Furuichi et al, 2010; Ligaba et al, 2013). OsALMT4 is expressed in roots and shoots and the OsALMT4 protein localizes to the plasma membrane. Overexpression of OsALMT4 can increase the efflux of malate in rice roots and the Al tolerance (Liu J et al, 2017).
One member of the natural resistance-associated macrophage proteins (Nramp), Nramp aluminum transporter 1 (Nrat1), is a plasma membrane-localized Al3+ transporter, which mediates the transport of Al3+ into root cells (Xia et al, 2010). Knockout of Nrat1 results in decreased Al uptake, while overexpression of Nrat1 leads to enhanced Al uptake (Xia et al, 2010; 2011). Natural variation of Nrat1 is present in rice germplasm. Famoso et al (2011) reported that Nrat1explains 40% of Al tolerance in an aus subpopulation. Xia et al (2014) also found that substitution of the chromosome segment containing Nrat1 from Koshihikari (Al tolerant) by that from Kasalath (Al sensitive) decreases Nrat1 expression and Al uptake and tolerance, but increases binding of Al to the cell wall. Expression of the tolerant OsNrat1 allele in yeast results in higher Al uptake than the sensitive allele and confers greater Al tolerance when expressed in transgenic Arabidopsis (Li et al, 2014). Therefore, different alleles of Nrat1 have different capacities in transporting Al into the root cells prior to final detoxification in the vacuoles.
The magnesium (Mg) transporter OsMGT1 is involved in Al tolerance through increasing Mg uptake (Chen Z C et al, 2012). In the presence of Al, Mg uptake is significantly enhanced in wild type rice, but not in OsMGT1 knockout lines. Further analysis of OsMGT1-mediated net Mg uptake in a kinetic experiment showed that Vmax increases two-fold in response to Al treatment, while Km is not altered (Chen Z C et al, 2012). These results indicate that Al has little effect on the affinity of OsMGT1 transporter for Mg, but it does induce the expression of OsMGT1 through regulation of ART1 (Yamaji et al, 2009), resulting in more OsMGT1 to participate in Mg transport. Thus, increasing Mg concentrations in the cytosol by OsMGT1 contributes to higher Al tolerance in rice (Chen and Ma, 2013).
OsALS1 encodes a half-size ABC transporter, a member of the TAP (transporter associated with antigen processing) sub-group regulated by ART1 (Yamaji et al, 2009). It is localized to the tonoplast. The expression of OsALS1 is rapidly and specifically induced by Al in the roots, but not by the other metals or low pH (Huang et al, 2012). Knockout of OsALS1 in three independent lines results in significantly increased sensitivity to Al, but does not affect the sensitivity to the other metals and low pH (Huang et al, 2012). Comparison of Al accumulation patterns between wild type and osals1 mutants showed that there is no significant difference in Al levels in the cell sap of root tips between wild type and the mutants, but the mutants accumulate more Al in the cytosol and nucleus than the wild type (Huang et al, 2012). These results indicate that OsALS1 is responsible for sequestration of Al into the vacuoles, which is required for internal detoxification of Al in rice.
Xia et al (2013) characterized a unique ART1- regulated gene, OsCDT3, which encodes a predicted peptide of only 53 amino acid residues located in plasma membrane. OsCDT3 is mainly expressed in the roots and its expression was specifically induced by Al exposure. Expression of OsCDT3 in yeast confers tolerance to Al, but not to Cd. Furthermore, OsCDT3 shows no transport activity for Al in yeast, but is able to directly bind Al in vitro. Knockdown of this gene results in decreased tolerance to Al, but does not affect the tolerance to Cd. The Al content decreases in the root residues including cell wall and the plasma membrane of the knockdown lines, but the Al content in the root cell sap increases compared with those of the wild type. Therefore, OsCDT3 anchoring to the plasma membrane may play a role in stopping entry of Al into the root cells by binding Al, therefore, contributing to high Al tolerance in rice (Xia et al, 2013).
The cell wall of root tip is the first barrier to prevent Al from entering the tissue, and the cell wall contains most of the Al in the whole root (Clarkson, 1967; Ma et al, 2004; Yang et al, 2008; Rangel et al, 2009). The primary root cell wall is mainly composed of cellulose, pectins and hemicelluloses (Sasidharan et al, 2011; Li et al, 2016). Pectin and hemicellulose are the main components, which carry the negative charges that can bind Al3+ in the cell wall, and modifications of their chemical structures or changes in their quantities significantly affect plant Al tolerance (Bosch and Hepler, 2005; Sasidharan et al, 2011; Yang et al, 2011; Li et al, 2016). There is a significant positive correlation between pectin content in the cell wall and Al accumulation in the root tip (Nagayama et al, 2019). Pectin-bound Al content is related to the pectin methylesterase (PME) regulated pectin methylesterifying degree (Yang et al, 2008; Rangel et al, 2010). The overexpression of OsPME14 in the Al-tolerant rice cultivar Nipponbare results in an increase in sensitivity to Al stress due to the significant increase in apoplastic Al content (Yang et al, 2013).
OsEXPA10 is an Al-inducible expansin gene involved in root cell elongation of rice (Che et al, 2016). The expression of OsEXPA10 in the roots is rapidly upregulated in response to Al stress (Tan et al, 2018). Knockout of OsEXPA10 results in a significant decrease in the cell elongation of the roots in the absence of Al. In the presence of Al, knockout of OsEXPA10 does not alter the Al sensitivity evaluated by relative root elongation, but the root cell wall of the knockout lines accumulates less Al compared to those of the wild type. These results indicate that OsEXPA10 expressed in the root tips is required for the root cell elongation, but that the contribution of this gene to high Al tolerance in rice is small (Che et al, 2016; Tan et al, 2018).
Membranes are the primary matrix for numerous physiological and biochemical activities, and plants easily change their membrane lipid compositions in response to environmental stress (Zhang et al, 2018). The galactolipids monogalactosyldiacylglycerol (MGDG) and digalactodiacylglycerol (DGDG) are major constituents of membranes in plants (Gaude et al, 2007; Maejima et al, 2014). One of the key enzymes for the biosynthesis of these galactolipids is MGDG synthase (MGD) (Wang et al, 2014). Compared with the wild type tobacco plants, the OsMGD-overexpression lines exhibit rapid regrowth of roots after removal of Al and less damage to membrane integrity and lipid peroxidation under Al stress. Meanwhile, the Al accumulation shows no difference between the wild type and transgenic plants. Lipid analysis showed that Al treatment dramatically decreases the content of MGDG and the ratio of MGDG to DGDG in the wild type plants, while it is unchanged in transgenic plants (Zhang M J et al, 2016). These results showed that the regulation of galactolipid biosynthesis can play an important role in maintaining membrane structure and function under Al stress.
A number of QTLs for Al tolerance have been identified by using different mapping populations and phenotyping methods. Four QTLs for Al tolerance have been identified on chromosomes 1, 3, 9 and 12 using relative root length in the mapping population of IR1552 × Azucena (Wu et al, 2000). Nguyen et al (2001) and Nguyen et al (2003) mapped five and ten QTLs related to Al tolerance using Chiembau × Omon269-65 and CT9993 × IR62266 populations, respectively. Xue et al (2007) mapped three QTLs for Al tolerance on chromosomes 1, 9 and 11 based on relative root elongation in a recombinant inbred population. Ma et al (2002) identified three QTLs for Al tolerance on chromosomes 1, 2 and 6 in the mapping population of Koshihikari × Kasalath with relative root elongation as a parameter. The Al transporter gene Nrat1 is responsible for the QTL previously detected on chromosome 2 by using chromosome segment substitution lines (Xia et al, 2014). DNA sequence variation in the Nrat1 coding and regulatory regions is associated with changes in Nrat1 expression and Nrat1 Al transport properties (Li et al, 2014).
Genome-wide association study (GWAS) is a powerful tool for unraveling the molecular basis for phenotypic diversity. GWAS of germplasms with rich variation could facilitate genetic mapping of multiple traits, which has become a complementary strategy to classical mapping with biparental synthetic recombinant populations (Huang and Han, 2014). Famoso et al (2011) firstly used GWAS on Al tolerance in rice and identified 48 regions associated with Al tolerance. Twenty-three QTLs for Al tolerance are obtained by GWAS of 150 rice accessions with relative root elongation (Zhang P et al, 2016). The multi-parent advanced generation inter-cross populations are genotyped using a 55 K rice SNP array and screened at the seedling stage for Al toxicity, and a total of 21 QTLs have been identified (Meng et al, 2017). Zhao et al (2018) used accessions with 67 511 SNPs for GWAS analysis, and identified 25 QTLs for Al tolerance on chromosomes 1, 4, 6, 7, 9 and 11. Tao et al (2018) performed an association mapping of Al toxicity tolerance using a core collection of 211 indica rice accessions with 700 K high quality SNP data, and 21 putative QTLs which affect shoot height, root length, root dry weight, shoot fresh weight, shoot dry weight and water content in shoot are identified at the seedling stage. GWAS performed in EMMAX for rice Al tolerance is carried out using 150 varieties of Ting’ s core collection, and 17 genes related to rice Al tolerance are identified (Zhang P et al, 2019).
Phytohormones such as auxin, ethylene, cytokinin, salycilic acid, jasmonic acid and abscisic acid have been reported to regulate the inhibition of Al-induced root growth (Kollmeier et al, 2000; Xue et al, 2008; Sun et al, 2010; Tamá s et al, 2012; Krishnamurthy and Rathinasabapathi, 2013; Yang et al, 2014, 2017). Liu et al (2016) showed that the inhibition of ethylene production by aminoethoxyvinylglycine (AVG, an ethylene synthesis inhibitor) treatment can suppress the Al-induced root swelling and ameliorate Al-induced root growth inhibition in rice, suggesting that increased production of ethylene is responsible for the root swelling under Al stress in rice. Pandey et al (2013) showed that exogenously added salicylic acid alleviates Al toxicity in rice seedlings by suppressing Al uptake, restoring root membrane integrity, reducing ROS level and ROS-induced oxidative damage and regulating the level of antioxidative enzyme activities. Saha et al (2019) studied the effect of abscisic acid on Al sensitivity through functioning of sub1A quantitative trait loci in rice cultivars, and found that abscisic acid is an important effector to modulate the Al tolerance pathway of rice cultivars through intrusion of sub1A. Recently, it has been reported that putrescine alleviates Al toxicity in rice by reducing Al content in cell wall in an ethylene-dependent manner (Zhu et al, 2019a).
Auxin plays rather multipurpose roles in Al resistance responses, including functioning as a signal molecule to respond to Al stress (Sun et al, 2010; Kochian et al, 2015), and regulating Al distribution within cell (Zhu et al, 2013; Yang et al, 2014; Bai et al, 2017; Liu X W et al, 2017). Auxin might be the connecting link that regulates the level of ROS and directs the role of ROS in oxidative stress (Iglesias et al, 2010; Krishnamurthy and Rathinasabapathi, 2013). OsAUX3 locates at the plasma membrane and functions as an auxin influx carrier affecting acropetal auxin transport (Wang et al, 2019). OsAUX3 is up-regulated in the root apex under Al stress, and osaux3 mutant is insensitive to Al treatments (Wang et al, 2019). Auxin concentrations, Al contents and Al-induced reactive oxygen species- mediated damage in osaux3 mutant under Al stress are lower than those in the wild type, indicating that OsAUX3 is involved in Al-induced inhibition of root growth (Wang et al, 2019).
Studies have shown that vesicular trafficking might be one of the earliest targets of Al toxicity in the root apexes (Amenó s et al, 2009; Krtková et al, 2012). Moreover, endocytosis might be involved in Al internalization (Illé š et al, 2006). Kollmeier et al (2000) found that Al, similarly to the inhibitors of polar auxin transport, such as 1-N-naphthyphthalamic acid (NPA) and 2, 3, 5-triiodobenzoic acid (TIBA), causes the inhibition of basipetal auxin transport, and thus inhibiting root growth. PIN-formed proteins, auxin efflux facilitators, direct the polar auxin transport and the asymmetric auxin distribution (Adamowski and Friml, 2015). These proteins rapidly and reversibly cycle between the plasma membrane and endosomes via vesicle trafficking (Kleine-Vehn et al, 2008; Adamowski and Friml, 2015). Endocytosis is an important internalization pathway for the intracellular uptake of portions of plasma membrane and extracellular cargos via pinching off vesicles from the plasma membrane (Samaj et al, 2004). Evidence from Arabidopsis further shows that this inhibitory effect of Al on auxin transport is associated with Al-blocked PIN2-mediated auxin polar transport (Sun et al, 2010), indicating that PIN2 may emerge as an Al-toxicity target of root apices (Baluska et al, 2010). OsPIN2 is located in plasma membrane and is an auxin efflux transporter. Overexpression of OsPIN2 can enhance auxin transport from shoot to root and auxin polar transport in roots (Chen Y N et al, 2012). Wu et al (2014) showed that OsPIN2 overexpression lines have significantly increased Al concentration in cell sap and reduced Al content in the cell wall in root apexes in rice compared to the wild type. By modulating PIN2-based auxin transport, auxin efflux and cell wall acidification, the OsPIN2 overexpression lines alleviate Al-induced cell rigidity in rice root apex (Wu et al, 2014). Wu et al (2015) further showed that endocytic vesicular trafficking may contribute to Al internalization, and that overexpressing OsPIN2 enhances rice Al tolerance via elevated endocytic vesicular trafficking and Al internalization.
Nitrogen (N) is the most abundant mineral nutrient element in plants, and plays an essential role in plant growth and development. Ammonium (NH4+) and nitrate (NO3-) are the two main inorganic N sources available for plant uptake (Xu et al, 2012; Chen et al, 2016, 2017). Zhao et al (2013) reported that Al3+ is beneficial to rice growth in medium containing NH4+, but harmful in medium containing NO3-. This suggests a synergistic effect between NH4+ and Al3+, but an antagonistic effect between NO3- and Al3+ (Zhao and Shen, 2013). The possible mechanism is that NH4+ decreases the cation-binding sites of the cell wall through direct NH4+ and indirect proton competition with Al ions (Zhao et al, 2009; Chen et al, 2010). Wang et al (2015) compared Al-binding capacity in the cell wall of the roots exposed to NH4+ and NO3- in two rice cultivars differing in Al tolerance, and found that the NH4+-reduced accumulation of Al3+ is a consequence of altered cell wall properties, which is triggered by pH decrease due to NH4+ uptake, rather than direct competition for the cell wall binding sites between Al3+ and NH4+.
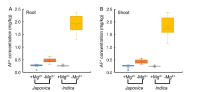
It has been reported that application of Mg in millimolar or micromolar concentrations can alleviate Al toxicity in a number of plant species (Tan et al, 1992; Kinraide et al, 2004; Watanabe and Okada, 2005; Bose et al, 2011, 2013; Chen Z C et al, 2012). Al3+ and Mg2+ compete for membrane transporters (Rengel and Robinson, 1989) and metal binding sites on enzymes (Grauer and Horst, 1992; Kinraide, 2003; Kinraide et al, 2004). Plants can reduce Al toxicity by enhancing Mg uptake. The involvement of Mg transporter in Al tolerance is first reported in yeast (ALR1 or ALR2) and Arabidopsis (AtMGT1) (MacDiarmid and Gardner, 1998; Deng et al, 2006). In wild type rice, Mg2+ uptake is enhanced and Mg2+ concentration in the cell sap of the root tips is also increased by Al (Chen Z C et al, 2012). OsMGT1 is expressed in both the roots and shoots in the absence of Al, but the expression is rapidly up-regulated by Al only in the roots (Chen Z C et al, 2012). When OsMGT1 is knocked out, Al tolerance is decreased, and the Al-induced inhibition of root elongation in the OsMGT1 knockout lines can be rescued by addition of 10 μ mol/L Mg2+ (Chen Z C et al, 2012). These findings indicated that up-regulation of OsMGT1 is required for conferring Al tolerance in rice by enhancing Mg2+ uptake (Chen Z C et al, 2012; Chen and Ma, 2013). In rice, japonica cultivars are usually more tolerant to Al than indica cultivars (Ma et al, 2002; Zhao et al, 2013). By analyzing the accumulation of Al3+ in 60 rice varieties (30 japonica and 30 indica) under Mg2+ treatment at the seedling stage, we found that the reduction of Mg2+ supply significantly increased the Al3+ uptake (Fig. 2). Mg2+ and Al3+ compete for membrane transporters, which can reduce the accumulation of Al3+ in plants (Rengel and Robinson, 1989; Kinraide et al, 2004; Bose et al, 2013). Compared with the supply of 1 mmol/L Mg2+, under the condition without Mg2+ supply, the Al3+ concentration of indica rice varieties was increased by six times, and that of japonica rice varieties increased by three times, both in the roots and shoots (Fig. 2). Zhao et al (2013) suggested that indica rice utilizes nitrate as nitrogen source more efficiently than japonica rice, and nitrate can promote the Al3+ accumulation in rice, resulting in the sensitivity of indica rice to aluminum toxicity. However, under the Mg2+ deficiency conditions, the reason why more Al3+ is accumulated in indica rice than in japonica rice is not clear, which needs further study.
Boron is an essential microelement for plant growth and is involved in alleviating Al toxicity (Ruiz et al, 2006; Yu et al, 2008; Li X W et al, 2018; Riaz et al, 2018; Yan et al, 2018). Zhu et al (2019b) found that 3 μ mol/L of boron pretreatment significantly enhances rice root elongation under Al toxicity conditions. Pretreatment with boron significantly decreases the deposition of Al in rice apoplasts, suppresses the synthesis of cell wall pectin, inhibits cell wall PME activity and its gene expression, and increases the expression of STAR1 and STAR2, which are responsible for reducing the Al content in the cell walls (Zhu et al, 2019b). Boron also increases the compartmentalization of Al from the cytoplasm to the vacuoles (Zhu et al, 2019b).
In low doses, hydrogen sulfide (H2S) functions as a signaling molecule in various physiological processes, such as improving seed germination under osmotic stress in wheat (Zhang et al, 2008, 2010), regulating the opening and closing of stomata in Arabidopsis (Liu et al, 2011; Jin et al, 2013), and improving resistance to salt stress in alfalfa (Wang et al, 2012), Al toxicity in barley (Chen et al, 2013), and oxidative stress in wheat under water-stress conditions (Shan et al, 2011). Zhu et al (2018) reported that pretreatment with 2 μ mol/L H2S significantly alleviates the inhibition of root elongation caused by Al toxicity in rice roots and Al contents in root tips are significantly reduced. NaHS pretreatment decreases the negative charge in cell walls by reducing the activity of pectin methylesterase and decreasing the pectin and hemicellulose contents in rice roots. Zhu et al (2018) inferred that H2S alleviates aluminum toxicity via decreasing apoplast and symplast Al contents in rice.
Great progresses have been made in understanding molecular mechanism of Al tolerance in rice during the last decades. The transcription factor ART1 plays a central role in Al tolerance in rice (Yamaji et al, 2009). The expression and localization of ART1 is unaffected by Al (Yamaji et al, 2009), therefore, there must be a process for activation of ART1. It will be interesting to identify ART1-interacting proteins in future. Only nine of the downstream genes of ART1 have been functionally characterized. Functional characterization of the remaining genes will extend our understanding on Al tolerance in rice.
At present, based on the study of Al tolerance at the rice seedling stage, some resistant varieties or materials have been screened, and several QTLs controlling Al tolerance have been identified by using molecular markers. However, there are still few studies on the fine mapping and cloning of the main QTLs. Understanding the genetic mechanism of rice Al tolerance is an important basis for rice Al tolerance breeding. Therefore, it is necessary to further study the screening, identification and functional analysis of the key QTLs for Al tolerance in rice. Transcriptome-wide association studies (TWAS) integrating with GWAS and gene expression datasets can be used to identify gene-trait associations (Wainberg et al, 2019). With the development of sequencing technology, TWAS can be used to identify new Al tolerance genes.
Marker-assisted selection (MAS) is a more acceptable method than the transgenic approaches for breeding. It is also clear that multiple genes for Al tolerance are needed to improve tolerance. Introduction of multiple Al-tolerance genes rather than a single gene will be more effective to improve Al tolerance and therefore increase rice production in acid soils (Ye and Smith, 2010; Guo and Ye, 2014).
Al toxicity is affected by the existence and status of key plant nutrition. For rice production in acid soils, proper application of Mg2+ fertilizer is an economic and effective way to improve rice tolerance to Al toxicity.
This research was financially supported by the National Natural Science Foundation of China (Grant No. 31902103), the Dapeng District Industry Development Special Funds (Grant No. KY20180218) and the Shenzhen Science and Technology Projects (Grant No. JSGG20160608160725473) in China.
(Managing Editor: Li Guan)
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
|
| [34] |
|
| [35] |
|
| [36] |
|
| [37] |
|
| [38] |
|
| [39] |
|
| [40] |
|
| [41] |
|
| [42] |
|
| [43] |
|
| [44] |
|
| [45] |
|
| [46] |
|
| [47] |
|
| [48] |
|
| [49] |
|
| [50] |
|
| [51] |
|
| [52] |
|
| [53] |
|
| [54] |
|
| [55] |
|
| [56] |
|
| [57] |
|
| [58] |
|
| [59] |
|
| [60] |
|
| [61] |
|
| [62] |
|
| [63] |
|
| [64] |
|
| [65] |
|
| [66] |
|
| [67] |
|
| [68] |
|
| [69] |
|
| [70] |
|
| [71] |
|
| [72] |
|
| [73] |
|
| [74] |
|
| [75] |
|
| [76] |
|
| [77] |
|
| [78] |
|
| [79] |
|
| [80] |
|
| [81] |
|
| [82] |
|
| [83] |
|
| [84] |
|
| [85] |
|
| [86] |
|
| [87] |
|
| [88] |
|
| [89] |
|
| [90] |
|
| [91] |
|
| [92] |
|
| [93] |
|
| [94] |
|
| [95] |
|
| [96] |
|
| [97] |
|
| [98] |
|
| [99] |
|
| [100] |
|
| [101] |
|
| [102] |
|
| [103] |
|
| [104] |
|
| [105] |
|
| [106] |
|
| [107] |
|
| [108] |
|
| [109] |
|
| [110] |
|
| [111] |
|
| [112] |
|
| [113] |
|
| [114] |
|
| [115] |
|
| [116] |
|
| [117] |
|
| [118] |
|
| [119] |
|
| [120] |
|
| [121] |
|
| [122] |
|
| [123] |
|
| [124] |
|
| [125] |
|
| [126] |
|
| [127] |
|
| [128] |
|
| [129] |
|
| [130] |
|
| [131] |
|
| [132] |
|
| [133] |
|
| [134] |
|
| [135] |
|
| [136] |
|
| [137] |
|
| [138] |
|
| [139] |
|
| [140] |
|
| [141] |
|
| [142] |
|
| [143] |
|
| [144] |
|
| [145] |
|
| [146] |
|
| [147] |
|
| [148] |
|