Plant lesion mimic mutants ( lmms) generally possess autoimmunity and hypersensitive response (HR)-like cell death in the absence of biotic or abiotic stress. They have attracted much attention because they are useful tools for deciphering the interaction between defense signaling and growth. Recent studies have identified more than 30 lmms involved in the plant immune response and cell death in rice. Genes underlying these lmms, coding for diverse types of proteins, mainly regulate transcription, protein translation and modification, vesicular trafficking and catalyzation of metabolism. Here, we presented an overview of the most recent advances on the study of lmmsin rice and proposed a perspective on potential utilization of LMM genes in agriculture.
Plants are sessile organisms that are constantly exposed to adverse environmental factors, including various pathogens. To survive from pathogen attack, plants have developed a robust innate immune system. During plant-pathogen interaction, many molecular events are activated to reprogram plants to fight against pathogens. Typical immune responses usually include production of reactive oxygen species (ROS), synthesis of antimicrobial metabolites, activation of pathogenesis-related (PR) genes, trafficking of related proteins or metabolites, protein modifications including phosphorylation and ubiquitination, changes in hormone levels and ion fluxes, and reinforcement of cell wall (Nurnberger et al, 2004; Tsuda and Katagiri, 2010; Henry et al, 2013). As a consequence, rapid cell death, characterized as hypersensitive response (HR), occurs in and around the infection site to restrict further invasion of pathogens, preventing disease development (Mur et al, 2008). However, activation of such an immune response usually delays plant growth (Lorrain et al, 2003).
Many mutants have been identified in maize, rice, Arabidopsis and other plants that exhibit spontaneous, HR-like cell death spots on their bodies even in the absence of pathogen infection, insect feeding, stress or mechanical damages (Hoisington et al, 1982; Lorrain et al, 2003; Huang et al, 2010). These mutants are usually named based on the cell death phenotypes, such as lesion mimic mutant (lmm), spotted leaf (spl), accelerated cell death (acd) and others (Lorrain et al, 2003; Cao et al, 2019). Here we employ more commonly used designation lmm to represent these types of mutants. These mutants usually carry characteristics of activated immune response, such as ROS burst, elevated levels of salicylic acid (SA), ethylene (ET) or jasmonic acid (JA), in the absence of pathogen attack.
It has been a long time since the first rice lmm mutant, sekiguchi lesion (sl), was identified and characterized (Sekiguchi, 1965). From then on, an increasing number of lmms from rice have been reported. Huang et al (2010) firstly reviewed progress of rice lmm research, mainly focusing on lmm sources and phenotypes, genetic models, and the six genes cloned at that time. During the past ten years, at least another 27 new rice lmms have been identified. These lmms usually present enhanced immune responses, elevated disease resistance and altered growth or developments (Supplemental Table 1), which provide new materials and tools for studying the crosstalk between immunity and growth.
| Supplemental Table 1. Summary of the thirty-three cloned LMM genes. |
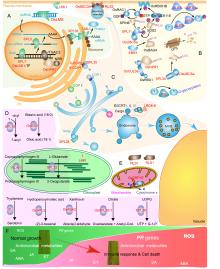
In this review, we mainly choose 33 rice LMM proteins for discussion. These LMMs mainly regulate four types of biological processes, including gene transcription and protein translation (Fig. 1-A), protein post-translational modifications (PTMs) (Fig. 1-B), vesicular trafficking (Fig. 1-C), and metabolic fluxes (Fig. 1-D). In addition, there are other regulators of immunity and cell death (Fig. 1-E). We discussed functions of these LMM genes and the crosstalk between immunity and growth, and provide thoughts for the implication of breeding rice with high yield and disease resistance.
Transcription is the first step for a gene to function in which the protein-coding DNA (gene) is transcribed by RNA polymerase II into a pre-mRNA. Pre-mRNA is subsequently spliced into a matured mRNA, which is used as a template to translate into a protein (Merchante et al, 2017). Transcription factors (TFs) including transcriptional activators and repressors play essential roles in the initiation of transcription and often determine the expression level of a gene (Merchante et al, 2017). The heat shock transcription factor SPOTTED LEAF (SPL) 7 represses immune response and cell death (Yamanouchi et al, 2002; Kojo et al, 2006). The spl7 mutant, containing a loss-of- function SPL7 protein with a base substitution in the heat stress transcription factor DNA-binding domain, displays HR-like cell death on leaves and accumulates more H2O2when incubated with elicitors prepared from cell wall extracts of the rice blast fungus Magnaporthe oryzae (Yamanouchi et al, 2002; Kojo et al, 2006). However, the detailed mechanism of how SPL7 regulates immunity and cell death is currently unknown. Zinc finger protein LESION SIMULATING DISEASE 1 (OsLSD1) functions together with certain TFs in nucleus to regulate rice immune response and cell death (Wang et al, 2005; Wu et al, 2014). Down-regulation of OsLSD1 expression by RNAi causes HR-like cell death. Interestingly, both elevation and inhibition of its mRNA levels lead to enhanced rice blast resistance, defining its unique and complex role in regulating immunity (Wang et al, 2005; Wu et al, 2014). NONEXPRESSOR OF PR GENES 1 (NPR1) is a transcriptional coactivator, a key protein controlling the establishment of systemic acquired resistance (SAR), an induced defense that spreads to the whole plant through long distance transport after exposure to various elicitors (Pajerowska-Mukhtar et al, 2013). Overexpression of OsNPR1/NPR1 Homolog 1(NH1) in rice can constitutively activate immune response and HR-like cell death, resulting in enhanced host resistance to both rice blast and bacterial blight diseases (Chern et al, 2005b; Yuan et al, 2007). The bacterial blight resistance mediated by OsNPR1 is weakened by NEGATIVE REGULATOR OF RESISTANCE (NRR). NRR interacts with OsNPR1 in the nucleus and suppresses the OsNPR1-mediated activation of defense genes (Chern et al, 2005a). Like Arabidopsis NPR1, OsNPR1 can also interact with TGA (TGACGTCA motif-binding factor) transcription factors to activate immune related genes (Chern et al, 2005b). While OsNPR1 enhances resistance to biotrophic pathogens, it reduces resistance to herbivores (Yuan et al, 2007). This may be due to the role of OsNPR1 in mediating an antagonistic crosstalk between SA- and JA-dependent pathways in rice (Yuan et al, 2007).
mRNA splicing is another regulatory step. SPL5 is a putative splicing factor 3b subunit 3 (SF3b3) which is thought to be involved in pre-mRNA processing (Chen et al, 2009, 2012). Deficiency of SPL5 activates a large number of genes involved in biotic defense responses and ROS metabolism, resulting in spontaneous cell death and elevated resistance to both blast and bacterial blight (Chen et al, 2009; Ge et al, 2015; Jin et al, 2015). lesion mimic and senescence(oslms) is another lmm with HR-like cell death that displays enhanced resistance to rice blast. OsLMS encodes a protein containing two double-stranded RNA binding motifs that is predicted to bind dsRNA (Undan et al, 2012). However, the substrates of OsLMS remain unidentified.
Eukaryotic translation elongation factor 1 alpha (eEF1A) has a pivotal role in protein synthesis by catalyzing GTP dependent binding of aminoacyl- tRNA to the acceptor site of the ribosome (Merchante et al, 2017). The eukaryotic translation termination factor 1 (eRF1) is also essential for protein synthesis by terminating and releasing the nascent polypeptide chains from the ribosome (Merchante et al, 2017). The rice eEF1A genes, SPL33/LMM5.1 andLMM5.4, and eRF1 gene LESION MIMIC LEAF 1 (LML1)/OsPELOTA are reported to regulate plant immunity and cell death. The spl33/lmm5and lml1/ospelotamutants display lesion mimic spots on leaves and enhance resistance to both rice blast and bacterial blight (Wang et al, 2017; Zhao et al, 2017; Qin et al, 2018; Zhang et al, 2018). Because the proteins are localized both on endoplasmic reticulum (ER) and plasma membrane, eRF1 LML1 may possess other functions in addition to its role in translation termination (Qin et al, 2018). Interestingly, SPL33/LMM5.1 and LMM5.4 are two eEF1As which share 97.4% amino acid sequence identity and are functionally redundant (Wang et al, 2017; Zhao et al, 2017). Due to natural variations in their promoters, the expression patterns of SPL33/LMM5.1 and LMM5.4 are different in japonicaand indica rice. In japonicarice, the expression of LMM5.4 is undetectable and its function is masked by SPL33/LMM5.1 (Zhao et al, 2017). Therefore, only the spl33/lmm5.1mutant, but not lmm5.4, displays the LMM phenotype in japonica rice and the function of the LMM5.4 locus is subsequently discovered through crossing lmm5.1 mutant with an indica rice (Wang et al, 2017; Zhao et al, 2017).
PTMs of proteins are a common phenomenon that widely exist in living organisms. Ubiquitination, phosphorylation and glycosylation are three well- known PTMs which regulate protein stability, activity, interaction and signal transduction (Yin et al, 2019). Their roles in immune response and cell death have also been uncovered in the study of rice lmms. SPL11 is an E3 ubiquitin ligase containing a U-box domain and an armadillo repeat domain. A single point mutation generates a premature stop codon in the SPL11 protein creating the spl11 mutant, which displays spontaneous cell death and enhanced resistance to fungal and bacterial pathogens (Zeng et al, 2004). SPL11 ubiquitinates SPL11 INTERACTING PROTEIN 6 (SPIN6), a Rho GTPase-activating protein, to tag it for degradation through the 26S proteasome-dependent pathway (Liu et al, 2015). Strikingly, SPIN6 interacts with the small GTPase OsRac1 which is a critical defense component in rice immune signaling. SPIN6 can catalyze the GTP- bound OsRac1 into the GDP-bound state, thus regulating immunity and cell death in rice (Liu et al, 2015). The monocot-specific receptor-like kinase SPL11 CELL DEATH SUPPRESSOR 2 (SDS2) is also suggested to be a substrate of SPL11. SDS2 phosphorylates SPL11, which in turn ubiquitinates SDS2 for degradation (Fan et al, 2018). SDS2 can also phosphorylate the receptor-like cytoplasmic kinases OsRLCK118 and the phosphorylated OsRLCK118 trans-phosphorylates OsRbohB to activate it, leading to ROS generation and defense gene expression (Fan et al, 2018). SPIN6 and SDS2, two positive regulators of immunity, are accumulated to higher levels in the spl11 mutant resulting in spontaneous HR-like cell death and enhanced resistance (Liu et al, 2015; Fan et al, 2018). Thus, the study of the spl11mutant revealed a complex signaling pathway linking many important cellular components. ENHANCED BLIGHT AND BLAST RESISTANCE 1 (EBR1), a RING-type E3 ubiquitin ligase, negatively regulates immunity and cell death in rice (You et al, 2016). The ebr1 mutant displays spontaneous cell death and enhanced disease resistance to rice blast and bacterial blight (You et al, 2016). In the ebr1 mutant, OsBAG4, a bcl2-associated athanogene family protein and a substrate of EBR1, accumulates. Accumulation of OsBAG4 triggers cell death and autoimmunity in rice (You et al, 2016). As a master regulator of SAR, the stability of NPR1 is also regulated by ubiquitination mediated by the Cullin3- based RING E3 ubiquitin ligase (CRL3), composed of Cullin3 (CUL3), RBX1 and BTB proteins. In rice, OsCUL3a is a CUL3 protein that mediates the ubiquitination and degradation of OsNPR1 (Liu et al, 2017). Loss-of-function of OsCUL3a activates OsNPR1- mediated immunity and cell death (Liu et al, 2017). Recently, the gene encoding a CUE (coupling of ubiquitin conjugation to ER degradation) domain containing protein SPL35 is cloned as another E3 ubiquitin ligase from a new lmm. The SPL35 protein is predominantly found in cytosol, ER and an unknown punctate compartment (Ma et al, 2019). SPL35 directly interacts with the E2 protein OsUBC5a, which also interacts with the SPL11 E3 ligase mentioned above (Bae and Kim, 2013). Knockdown of SPL35 or OsUBC5a leads to an LMM phenotype in rice, suggesting that the SPL35-OsUBC5a module is involved in immunity and cell death regulation. SPL35 is also found to interact with monoubiquitin through its CUE domain (Ma et al, 2019). However, the substrate of the SPL35-OsUBC5a module is currently unknown. Moreover, overexpression of SPL35 also results in enhanced immune response and cell death, indicating a much complex network governed by SPL35 (Ma et al, 2019).
Many protein kinases are responsible for rice lmms, indicating that protein phosphorylation is main step regulating immunity and cell death in rice. A mitogen- activated protein kinase kinase kinase (MAPKKK), named SPL3/ENHANCED DISEASE RESISTANCE 1 (OsEDR1)/ACCELERATED CELL DEATH AND RESISTANCE 1 (OsACDR1), is reported to regulate HR-like cell death formation (Kim et al, 2009; Shen et al, 2011; Wang S H et al, 2015). Overexpression of OsACDR1 in rice confers spontaneous HR-like cell death and enhanced disease resistance (Kim et al, 2009). Interestingly, loss-of-function of SPL3/OsEDR1/ OsACDR1 also activates immunity and cell death in rice (Shen et al, 2011; Wang S H et al, 2015). The LMM phenotype of spl3/osedr1 is likely resulting from the altered levels of hormones such as SA, JA, ET and abscisic acid (ABA) (Shen et al, 2011; Wang S H et al, 2015). However, how this MAPKKK regulates immunity and cell death through hormones remains unclear. SPOTTED LEAF SHEATH (SLES) is the second MAPKKK cloned from a rice lmm. The sles displays lesion mimic spots mainly on leaf sheaths, resulting in early leaf senescence and enhanced defense response (Lee et al, 2018). Two plasma membrane protein kinases, PTO-INTERACTING PROTEIN 1a (OsPTI1a) and APOPTOSIS LEAF AND SHEATH 1 (ALS1)/LIGHT INDUCED LESION MIMIC MUTANT 1 (LIL1) are implicated in the pathways of cell death and autoimmunity. OsPTI1a localizes to plasma membrane through its N-terminal palmitoylation (Matsui et al, 2015). Proper complex formation mediated by OsPTI1a at the plasma membrane is required for the negative regulation of plant immune responses and cell death in rice (Matsui et al, 2015). The immune response and cell death regulated by OsPTI1a is dependent on RAR1 which is a main regulator of R proteins involved in disease resistance (Takahashi et al, 2007). The cysteine-rich receptor like kinase ALS1/LIL1 is identified from two semi-dominant ricelmms (Zhou et al, 2017; Du et al, 2019). The mutants display HR-like cell death spots on their leaf blades and leaf sheaths and constitutively activated defense response, resembling SA-, JA- and OsNPR1/NH1-mediated defense responses in rice (Zhou et al, 2017; Du et al, 2019). Overexpression of OsNPR1/NH1 readily leads to a lesion mimic phenotype and enhanced disease resistance, which is inducible by application of SAR inducer benzothiadiazole (BTH); deletion of two cysteine-rich receptor-like kinases (CRK6 and CRK10) in a rice mutant (snim1) blocks this OsNPR1/NH1-mediated, BTH-inducible lesion mimic phenotype and enhances disease resistance, showing the involvement of receptor-like kinases in OsNPR1/NH1 function in rice (Chern et al, 2016).
In addition, glycosylation is also suggested to regulate rice immunity and cell death. The PREMATURE SENESCENCE LEAF (PSL) gene encodes a putative core 2/I branching beta-1, 6-N-acetylglucosaminyl transferase that is predicted to function in protein glycosylation. In the psl mutant, the O-glycosylation is impaired while the ET-related metabolic process is activated. The psl mutant exhibits spontaneous cell death and autoimmunity (Ke et al, 2019). This discovery highlights the importance of protein O-glycosylation in cell death and immunity.
Plant cells usually contain many membrane-bounded compartments that interact with the cellular environment to regulate various aspects of plant growth (Paul et al, 2014). Vesicular transport pathways play essential roles in cargo (usually proteins, lipids and other biomolecules) trafficking between these compartments. Many types of vesicles have been identified to participate in vesicular transport in the endocytic and secretory pathways (Paul et al, 2014). The coat protein complex I (COPI), COPII and clathrin-coated vesicles and their adaptor complexes are important for sorting and directing the transportation of cargos (Paul et al, 2014; Gomez-Navarro and Miller, 2016). The multivesicular bodies (MVBs) and secretory vesicles are also important for cargo transportation. Here, we mainly focused on trafficking pathways and components which are involved in autoimmunity and spontaneous cell death in rice. For more details on how these vesicles are assembled and transported, readers are referred to other reviews (McMahon and Mills, 2004; Cui et al, 2016; Mei and Guo, 2018).
COPI-coated vesicles are involved in retrograde transport from the Golgi apparatus to ER (Gomez- Navarro and Miller, 2016). In rice, the coatomer subunits δ -COP1 and δ -COP2 interact directly with SPL35, a CUE domain-containing protein discussed above. Knockdown of δ -COP1 or δ -COP2 gene leads to constitutive immunity activation and HR-like cell death (Ma et al, 2019). SPL35 protein is found to distribute in some cytosolic punctate compartments which are likely the COPI-coated vesicles, providing evidence that the COPI trafficking pathway is involved in the regulation of HR-like cell death and immunity in rice (Ma et al, 2019). The clathrin- associated adaptor protein complex 1, medium subunit μ 1 (AP1M1) protein SPL28, is localized to the Golgi apparatus and likely involved in the regulation of clathrin-associated adaptor protein complex 1 (AP-1)- mediated membrane-cytoplasm recycling during transport vesicle formation and vesicle uncoating in rice (Qiao et al, 2010). The spl28 mutant accumulates antimicrobial compounds and activates defense response to rice blast and bacterial blight pathogens, suggesting that the clathrin-coated vesicle pathway is also involved in HR-like cell death and immunity in rice (Qiao et al, 2010). The MVB-mediated trafficking pathway is regulated by an AAA type ATPase LRD6-6. Defect in its ATPase activity leads to autoimmunity and HR-like cell death (Zhu et al, 2016). LRD6-6 interacts with the endosomal sorting complexes required for transport of (ESCRT)-III components OsSNF7 and OsVPS2, and co-localizes with the MVB marker protein RabF1/ARA6. Moreover, the transportation of soluble vacuolar carboxypeptidase Y from ER to vacuoles, which is mediated by MVBs, is blocked in the lrd6-6mutant (Zhu et al, 2016). Thus, activated immunity and enhanced cell death in lrd6-6 is due to the inhibition of trafficking pathway mediated by MVBs (Zhu et al, 2016). The exocyst is an evolutionarily conserved protein complex composed of SEC3, SEC5, SEC6, SEC8, SEC10, SEC15, EXO70 and EXO84 that are involved in exocytosis (Mei and Guo, 2018). Rice OsSEC3A binds to phosphatidylinositol and interacts with the SNAP25-type t-SNARE protein OsSNAP32, which is involved in rice blast resistance (Ma et al, 2018). Knockout of OsSEC3a causes HR-like cell death and enhanced defense responses. OsSEC3A has a punctate distribution on the plasma membrane which matches its role (Ma et al, 2018). Loss-of-function of another exocyst complex subunit OsEXO70A1 inrapid leaf senescence 2 (rls2) also confers an LMM phenotype. Although the immune response of the rls2 mutant is not surveyed, the secretory pathway is thus also believed to be involved in regulating rice immunity and cell death based on the phenotype (Tu et al, 2015).
Living organisms metabolize nutrients as sources of energy and building blocks for proteins, lipids, nucleic acids and carbohydrates to survive (Green et al, 2014). Dysfunction of several metabolic fluxes can cause accumulation of intermediate metabolites, leading to autoimmunity and cell death. Currently, at least nine key enzymes involved in different metabolic pathways have been identified in the study of rice lmms that confer autoimmunity and cell death. SPL18, an acyl transferase, likely contributes to disease resistance by potentiating disease-resistance signaling through its enzymatic activity (Mori et al, 2007). It may also act by producing phytoalexin-like secondary products. Therefore, when Spl18 mRNA accumulates to high levels, the immune response is activated and cell death occurs (Mori et al, 2007). Rice SUPPRESSOR OF SA INSENSITIVE 2 (OsSSI2) is a fatty acid desaturase that catalyzes the conversion of stearic acid (18:0) to oleic acid (18 : 1). Knockdown of OsSsi2 accumulates stearic acid (18 : 0), thus increases endogenous free SA levels and induces SA-responsive genes, leading to enhanced disease resistance and spontaneous HR-like cell death (Jiang et al, 2009). A novel defense pathway was also discovered in the study of rice lmm sekiguchi lesion (sl) (Fujiwara et al, 2010). SL encodes a CYP71P1 protein which exhibits tryptamine 5-hydroxylase enzymatic activity and catalyzes the conversion of tryptamine to serotonin (Fujiwara et al, 2010). Rice sl mutant exhibits unique orange-colored lesions and accumulates tryptamine. Exogenous application of serotonin is capable of inducing defense responses and cell death mediated by the RacGTPase pathway and the Gα subunit of the heterotrimeric G protein (Fujiwara et al, 2010). The rice lesion initiation 1 (rlin1)/leaf lesion mimic mutant 1 (llm1) exhibits cell death on leaf and enhanced disease resistance to bacterial blight pathogen (Wang J et al, 2015). Rlin1 encodes a putative coproporphyrinogen III oxidase functioning in the tetrapyrrole biosynthesis pathway, suggesting that this pathway is involved in rice immunity and cell death (Sun et al, 2011; Wang J et al, 2015). Hydroperoxylinoleic acid is catabolized by hydroperoxide lyase (HPL) to form (Z)-3-hexenal or by allene oxide synthase to form JA in the oxylipin pathway (Liu X Q et al, 2012). Loss-of-function of a rice HPL, OsHPL3/CYP74B2, leads to accumulation of JA but reduction of green leaf volatiles in the hpl3/cea62 mutant (Liu X Q et al, 2012; Tong et al, 2012). Thus, OsHPL3 modulates rice defense responses against different invaders, including pathogens and insects through the oxylipin pathway (Liu X Q et al, 2012; Tong et al, 2012). The UDP-N-acetylglucosamine pyrophosphorylase 1 (UAP1) SPL29 catalyzes the decomposition of uridine 5′ -diphosphoglucose-glucose (UDPG), an important signaling molecule involved in immunity (Xiao et al, 2018). Loss-of-function of Spl29can accumulate excessive UDPG, leading to a ROS burst, immune response and cell death (Wang Z H et al, 2015; Xiao et al, 2018). AnotherSpl gene, Spl32, encodes a chloroplast-localized ferredoxin dependent glutamate synthase, which is a key enzyme in the process of inorganic nitrogen assimilation (Sun et al, 2017). The spl32 mutant displays spontaneous cell death and elevated disease resistance which likely results from reduction of photorespiration rate and burst of ROS (Sun et al, 2017). The lmm9150 mutant exhibits spontaneous cell death and enhanced resistance to rice bacterial and blast diseases. OsABA2, a xanthoxin dehydrogenase involved in ABA biosynthesis pathway, is responsible for this phenotype (Liao et al, 2018). Consistent with OsABA2 function, ABA level in the lmm9150 mutant is significantly reduced (Liao et al, 2018). This discovery directly links biosynthesis of ABA, which causes senescence, with immunity and cell death. Recently, an ATP-citrate lyase (ACL), a subunit protein, SPL30, is reported to regulate immunity and cell death in rice (Ruan et al, 2018). The lmm spl30-1 harbors A-to-T substitution that converts an asparagine to tyrosine (N343Y), which causes significant degradation of SPL30N343Y in a ubiquitin- dependent manner and reduces the ACL enzymatic activity in planta (Ruan et al, 2018). Further suppressor screens revealed that SPL30 is epistatic to SL in the pathogen response pathway, linking Spl30 to a previously known serotonin activated immune pathway (Ruan et al, 2018).
Apart from the above pathways, other types of regulators controlling immunity and cell death in rice have been characterized. Resistance (R) proteins are key components in mediating defense responses. The semi-dominant mutants, necrotic leaf sheath 1 (nls1- 1D) and nls1-2D, are found to display spontaneous cell death on leaf sheaths and constitutively activate defense responses (Tang et al, 2011). The NLS1 gene encodes a typical coiled-coil, nucleotide binding and leucine rich repeat domain (CC-NB-LRR)-containing R protein. Both nls1-1D and nls1-2D mutations cause constitutive auto-activation of the NLS1 R protein leading to a SA- and NPR1-independent defense response (Tang et al, 2011). The conserved nucleotide-binding (NB) domain has been also referred to as NB-ARC (nucleotide-binding, Apaf-1, R proteins, and Ced-4) (van der Biezen and Jones, 1998). RAPID LEAF SENESCENCE 1 (RLS1), a NB-ARC domain- containing protein, is also found to regulate cell death in rice. The rls1 mutant displays yellow brown lesions and accelerated leaf senescence while immune responses in rls1 is not studied (Jiao et al, 2012).
Cytochrome c plays an important role in mitochondria governing apoptosis in animals (Hoeberichts and Woltering, 2003). However, how cytochrome c is released from the mitochondria to cytosol remains poorly understood. In the study of a rice lmm mutant dj-lm, the mitochondria-localized GTPase DYNAMIN- RELATED PROTEIN 1E (OsDRP1E) is found to negatively regulate cell death and immunity by controlling mitochondrial structure and cytochrome c release (Li Z Q et al, 2017). Mitochondrial morphology is changed in the dj-lm mutant and concentration of cytoplasmic cytochrome c is increased. As a result, the dj-lm mutant exhibits spontaneous cell death and enhanced resistance to fungal and bacterial pathogens (Li Z Q et al, 2017). This discovery advances our knowledge on the relationship among cytochrome c release, immunity and cell death in rice.
The LMM phenotype is primarily characterized by the presence of brown-spots on rice leaf blades or leaf sheaths. These brown-spots are reminiscent of HR which is regarded as a form of rapid programmed cell death that occurs in host cells during plant-pathogen interaction (Coll et al, 2011). Molecular events such as burst of ROS, accumulation of PR mRNAs and elevation of hormone levels are accompanied with HR (Coll et al, 2011). Indeed, these events autonomously happen in lmms, priming immune response resulting in HR-like cell death phenotypes. Almost all lmms accumulate elevated levels of ROS and transcripts of PR genes. A majority of the mutants, such as oshpl3, ebr1, nls1, spl29 and lmm9150, also include imbalanced hormone levels, including SA, JA, ET and ABA (Tang et al, 2011; Tong et al, 2012; You et al, 2016; Liao et al, 2018; Xiao et al, 2018). The antimicrobial metabolites phytoalexins, serotonin and phenolic compounds also accumulate to high concentrations in some of the lmms, like spl28, lrd6-6, spl5, sl andspl3 (Fujiwara et al, 2010; Qiao et al, 2010; Jin et al, 2015; Wang S H et al, 2015; Zhu et al, 2016). The deposition of callose and increase of cell wall component lignin are also detected in these lmms (Qiao et al, 2010; Zhu et al, 2016). All these are hallmarks of a typical immune response in plants.
Obviously, although the types of LMM genes and their pathways are highly diverse, their downstream molecular events triggering the immune response and cell death are conserved (Fig. 1-F). Thus, lmmsare of great significance for deciphering upstream regulation of defense signaling pathways.
Normal growth can be suppressed on activation of the immune response, resulting in growth delay and yield penalties in plants (Yin et al, 2019). Thus, high yield with enhanced resistance is a highly desirable trait but also a major challenge in crop breeding.
The sustained activation of immune response results in cell death and huge influences on many aspects of plant growth in rice lmms (Supplemental Table 1). It is very likely that adverse effects on normal growth of lmms may result from the altered photosynthetic system caused by the immune response and cell death. This preliminary conclusion is based on the following observations: (1) Many lmms, such as rls1, spl32 and spl29, carrying weak growth are reported to display abnormal degradation of chloroplasts and reduced photosynthetic capacity (Jiao et al, 2012; Sun et al, 2017; Xiao et al, 2018). (2) Mutants like spl3, spl4 and lmm9150 show stay-green or delayed leaf senescence phenotype which usually represents a longer durations of photosynthetic activity and growth closer to the wild type at certain stages even though some aspects of agronomic traits are still affected (Wang S H et al, 2015; Liao et al, 2018; Song et al, 2018). (3) More interestingly, overexpression of OsLSD1 promotes chlorophyll b accumulation, leading to dark-green-colored leaves, enhanced disease resistance without cell death, and accelerated differentiation of callus (Wang et al, 2005).
Several studies of lmms have gained some insights into molecular mechanisms governing the balance between immunity and growth. The E3 ubiquitin ligase SPL11 negatively regulates programmed cell death and disease resistance (Liu J L et al, 2012). And, SPL11 interacts with mono-ubiquitinates SPIN1, a flowering time regulator involved in Heading date3a (Hd3a) pathway, to cause delayed flowering in spl11mutant under long-day conditions (Vega-Sanchez et al, 2008). While over accumulated in leaves, OsLSD1increases chlorophyll b content and disease resistance without visible cell death (Wang et al, 2005). Moreover, OsLSD1 can interact with OsbZIP58 in seed to activate gibberellin biosynthesis gene OsKO2 and to downregulate the SOD1-encoding gene to modulate seed germination (Wu et al, 2014). In order to facilitate utilization of LMM genes, attentions should be paid to the roles of LMM genes on regulation of growth in the future.
Several genes such as IPA1, Bsr-d1, Bsr-k1 and the Pigm pairs have been characterized to play important roles in balancing yield and resistance in rice (Deng et al, 2017; Li W T et al, 2017; Wang et al, 2018; Zhou et al, 2018). These discoveries provide precious gene resources which are useful for crop breeding. Unlike the resistance provided by these genes, mutations in LMM genes always result in sustained and overactive activation of immune responses which causes HR-like cell death and adversely impacts plant growth. Thus, it is difficult to directly apply those LMM genes to breeding application, especially for the recessive genes. However, the studies on these lmms provide insights to understand the crosstalk between immunity and growth. For examples, in-depth study on LMM gene SPL11 has strengthened our understanding on how a E3 ligase regulates plant immunity and flowering (Vega- Sanchez et al, 2008; Liu et al, 2015; Fan et al, 2018). Due to activation of cell death, the photosynthetic capacity of some lmms is reduced which might inhibit plant development (Jiao et al, 2012; Sun et al, 2017; Xiao et al, 2018).
It is common that enhanced resistance obtained is often associated with substantial penalties to fitness. It is not only true to LMM genes, but also to other resistance genes. However, previous studies showed that we can overcome this problem by driving these defense genes under control of pathogen specific, inducible promoters. TBF1 is identified as an important transcription factor controlling the growth-to-defense switch upon immune induction. Translation of TBF1 is normally inhibited by two upstream open reading frames within the 5′ leader sequence (Pajerowska- Mukhtar et al, 2012). Using the TBF1-cassette as a promoter to drive NPR1 has been shown to enhance rice broad-spectrum disease resistance with minimal adverse effects on plant growth and development (Xu et al, 2017). The promoter of OsHEN1 contains a binding site of TAL9, which is a transcriptional activator- like effector secreted by the type III secretion system (Moscou and Bogdanove, 2009). The expression of OsHEN1 is specifically induced by pathogens containing TAL9a. Using the OsHEN1promoter to drive IPA1 gene has achieved high yield and enhanced resistance in rice (Liu et al, 2019). Promoters, like the TBF1- cassette and the OsHEN1promoter, are ubiquitous in plant genomes. Identification and isolation of more such promoters would provide useful tools.
This study was supported by the National Natural Science Foundation of China (Grant Nos. 31701779 and 31922066), the Applied Basic Research Programs of Science and Technology Department from Sichuan Province (Grant No. 2019YJ0432) and China Postdoctoral Science Foundation (Grant No. 2017M612984).
The following material is available in the online version of this article at http://www.sciencedirect.com/science/ journal/16726308; http://www.ricescience.org.
Supplemental Table 1. Summary of 23 cloned LMM genes.
Chen Xifeng, Hao Liang, Pan Jianwei, Zheng Xixi, Jiang Guanghuai, Jin Yang, Gu Zhimin, Qian Qian, Zhai Wenxue, Ma Bojun 2012. SPL5, a cell death and defense-related gene, encodes a putative splicing factor 3b subunit 3 SF3b3) in rice. Molecular breeding, 302): 939-949.
Chern M., Fitzgerald H. A., Canlas P. E., Navarre D. A., Ronald P. C. 2005. Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Mol Plant Microbe Interact, 186: 511-520.
Du D., Liu M., Xing Y., Chen X., Zhang Y., Zhu M., Lu X., Zhang Q., Ling Y., Sang X., Li Y., Zhang C., He G. 2019. Semi-dominant mutation in the cysteine-rich receptor-like kinase gene, ALS1, conducts constitutive defence response in rice. Plant Biol Stuttg, 211: 25-34.
Fan J., Bai P., Ning Y., Wang J., Shi X., Xiong Y., Zhang K., He F., Zhang C., Wang R., Meng X., Zhou J., Wang M., Shirsekar G., Park C. H., Bellizzi M., Liu W., Jeon J. S., Xia Y., Shan L., Wang G. L. 2018. The Monocot-Specific Receptor-like Kinase SDS2 Controls Cell Death and Immunity in Rice. Cell Host Microbe, 234: 498-510 e495.
Fekih R., Tamiru M., Kanzaki H., Abe A., Yoshida K., Kanzaki E., Saitoh H., Takagi H., Natsume S., Undan J. R., Undan J., Terauchi R. 2015. The rice Oryza sativa L. LESION MIMIC RESEMBLING, which encodes an AAA-type ATPase, is implicated in defense response. Mol Genet Genomics, 2902: 611-622.
Fujiwara T., Maisonneuve S., Isshiki M., Mizutani M., Chen L., Wong H. L., Kawasaki T., Shimamoto K. 2010. Sekiguchi lesion gene encodes a cytochrome P450 monooxygenase that catalyzes conversion of tryptamine to serotonin in rice. J Biol Chem, 28515: 11308-11313.
Ge Chang-wei, Zhi-guo E, Pan Jiang-jie, Jiang Hua, Zhang Xiao-qin, Zeng Da-li, Dong Guo-jun, Hu Jiang, Xue Da-wei 2015. Map-based cloning of a spotted-leaf mutant gene OsSL5 in Japonica rice. Plant growth regulation, 753: 595-603.
Jiang C. J., Shimono M., Maeda S., Inoue H., Mori M., Hasegawa M., Sugano S., Takatsuji H. 2009. Suppression of the rice fatty-acid desaturase gene OsSSI2 enhances resistance to blast and leaf blight diseases in rice. Mol Plant Microbe Interact, 227: 820-829.
Jiao B. B., Wang J. J., Zhu X. D., Zeng L. J., Li Q., He Z. H. 2012. A novel protein RLS1 with NB-ARM domains is involved in chloroplast degradation during leaf senescence in rice. Mol Plant, 51: 205-217.
Jin B., Zhou X., Jiang B., Gu Z., Zhang P., Qian Q., Chen X., Ma B. 2015. Transcriptome profiling of the spl5 mutant reveals that SPL5 has a negative role in the biosynthesis of serotonin for rice disease resistance. Rice N Y, 8: 18.
Ke S., Liu S., Luan X., Xie X. M., Hsieh T. F., Zhang X. Q. 2019. Mutation in a putative glycosyltransferase-like gene causes programmed cell death and early leaf senescence in rice. Rice N Y, 121: 7.
Kim J. A., Cho K., Singh R., Jung Y. H., Jeong S. H., Kim S. H., Lee J. E., Cho Y. S., Agrawal G. K., Rakwal R., Tamogami S., Kersten B., Jeon J. S., An G., Jwa N. S. 2009. Rice OsACDR1 Oryza sativa accelerated cell death and resistance 1 is a potential positive regulator of fungal disease resistance. Mol Cells, 285: 431-439.
Kojo K., Yaeno T., Kusumi K., Matsumura H., Fujisawa S., Terauchi R., Iba K. 2006. Regulatory mechanisms of ROI generation are affected by rice spl mutations. Plant Cell Physiol, 478: 1035-1044.
Lee D., Lee G., Kim B., Jang S., Lee Y., Yu Y., Seo J., Kim S., Lee Y. H., Lee J., Kim S., Koh H. J. 2018. Identification of a Spotted Leaf Sheath Gene Involved in Early Senescence and Defense Response in Rice. Front Plant Sci, 9: 1274.
Li Z., Ding B., Zhou X., Wang G. L. 2017. The Rice Dynamin-Related Protein OsDRP1E Negatively Regulates Programmed Cell Death by Controlling the Release of Cytochrome c from Mitochondria. PLoS Pathog, 131: e1006157.
Liao Y., Bai Q., Xu P., Wu T., Guo D., Peng Y., Zhang H., Deng X., Chen X., Luo M., Ali A., Wang W., Wu X. 2018. Mutation in Rice Abscisic Acid2 Results in Cell Death, Enhanced Disease-Resistance, Altered Seed Dormancy and Development. Front Plant Sci, 9: 405.
Liu J., Li W., Ning Y., Shirsekar G., Cai Y., Wang X., Dai L., Wang Z., Liu W., Wang G. L. 2012a. The U-Box E3 ligase SPL11/PUB13 is a convergence point of defense and flowering signaling in plants. Plant Physiol, 1601: 28-37.
Liu J., Park C. H., He F., Nagano M., Wang M., Bellizzi M., Zhang K., Zeng X., Liu W., Ning Y., Kawano Y., Wang G. L. 2015. The RhoGAP SPIN6 associates with SPL11 and OsRac1 and negatively regulates programmed cell death and innate immunity in rice. PLoS Pathog, 112: e1004629.
Liu Q., Ning Y., Zhang Y., Yu N., Zhao C., Zhan X., Wu W., Chen D., Wei X., Wang G. L., Cheng S., Cao L. 2017. OsCUL3a Negatively Regulates Cell Death and Immunity by Degrading OsNPR1 in Rice. Plant Cell, 292: 345-359.
Liu X., Li F., Tang J., Wang W., Zhang F., Wang G., Chu J., Yan C., Wang T., Chu C., Li C. 2012b. Activation of the jasmonic acid pathway by depletion of the hydroperoxide lyase OsHPL3 reveals crosstalk between the HPL and AOS branches of the oxylipin pathway in rice. PLoS One, 711: e50089.
Ma J., Chen J., Wang M., Ren Y., Wang S., Lei C., Cheng Z., Sodmergen 2018. Disruption of OsSEC3A increases the content of salicylic acid and induces plant defense responses in rice. J Exp Bot, 695: 1051-1064.
Ma J., Wang Y., Ma X., Meng L., Jing R., Wang F., Wang S., Cheng Z., Zhang X., Jiang L., Wang J., Wang J., Zhao Z., Guo X., Lin Q., Wu F., Zhu S., Wu C., Ren Y., Lei C., Zhai H., Wan J. 2019. Disruption of gene SPL35, encoding a novel CUE domain-containing protein, leads to cell death and enhanced disease response in rice. Plant Biotechnol J.
Matsui H., Takahashi A., Hirochika H. 2015. Rice immune regulator, OsPti1a, is specifically phosphorylated at the plasma membrane. Plant Signal Behav, 103: e991569.
Mori M., Tomita C., Sugimoto K., Hasegawa M., Hayashi N., Dubouzet J. G., Ochiai H., Sekimoto H., Hirochika H., Kikuchi S. 2007. Isolation and molecular characterization of a Spotted leaf 18 mutant by modified activation-tagging in rice. Plant Mol Biol, 636: 847-860.
Qiao Y., Jiang W., Lee J., Park B., Choi M. S., Piao R., Woo M. O., Roh J. H., Han L., Paek N. C., Seo H. S., Koh H. J. 2010. SPL28 encodes a clathrin-associated adaptor protein complex 1, medium subunit micro 1 AP1M1 and is responsible for spotted leaf and early senescence in rice Oryza sativa. New Phytol, 1851: 258-274.
Qin P., Fan S., Deng L., Zhong G., Zhang S., Li M., Chen W., Wang G., Tu B., Wang Y., Chen X., Ma B., Li S. 2018. LML1, Encoding a Conserved Eukaryotic Release Factor 1 Protein, Regulates Cell Death and Pathogen Resistance by Forming a Conserved Complex with SPL33 in Rice. Plant Cell Physiol, 595: 887-902.
Ruan B., Hua Z., Zhao J., Zhang B., Ren D., Liu C., Yang S., Zhang A., Jiang H., Yu H., Hu J., Zhu L., Chen G., Shen L., Dong G., Zhang G., Zeng D., Guo L., Qian Q., Gao Z. 2018. OsACL-A2 negatively regulates cell death and disease resistance in rice. Plant Biotechnol J.
Shen X., Liu H., Yuan B., Li X., Xu C., Wang S. 2011. OsEDR1 negatively regulates rice bacterial resistance via activation of ethylene biosynthesis. Plant Cell Environ, 342: 179-191.
Song G., Kwon C. T., Kim S. H., Shim Y., Lim C., Koh H. J., An G., Kang K., Paek N. C. 2018. The Rice SPOTTED LEAF4 SPL4 Encodes a Plant Spastin That Inhibits ROS Accumulation in Leaf Development and Functions in Leaf Senescence. Front Plant Sci, 9: 1925.
Sun C., Liu L., Tang J., Lin A., Zhang F., Fang J., Zhang G., Chu C. 2011. RLIN1, encoding a putative coproporphyrinogen III oxidase, is involved in lesion initiation in rice. J Genet Genomics, 381: 29-37.
Sun L., Wang Y., Liu L. L., Wang C., Gan T., Zhang Z., Wang Y., Wang D., Niu M., Long W., Li X., Zheng M., Jiang L., Wan J. 2017. Isolation and characterization of a spotted leaf 32 mutant with early leaf senescence and enhanced defense response in rice. Sci Rep, 7: 41846.
Takahashi A., Agrawal G. K., Yamazaki M., Onosato K., Miyao A., Kawasaki T., Shimamoto K., Hirochika H. 2007. Rice Pti1a negatively regulates RAR1-dependent defense responses. Plant Cell, 199: 2940-2951.
Tang J., Zhu X., Wang Y., Liu L., Xu B., Li F., Fang J., Chu C. 2011. Semi-dominant mutations in the CC-NB-LRR-type R gene, NLS1, lead to constitutive activation of defense responses in rice. Plant J, 666: 996-1007.
Tong X., Qi J., Zhu X., Mao B., Zeng L., Wang B., Li Q., Zhou G., Xu X., Lou Y., He Z. 2012. The rice hydroperoxide lyase OsHPL3 functions in defense responses by modulating the oxylipin pathway. Plant J, 715: 763-775.
Tu B., Hu L., Chen W., Li T., Hu B., Zheng L., Lv Z., You S., Wang Y., Ma B., Chen X., Qin P., Li S. 2015. Disruption of OsEXO70A1 Causes Irregular Vascular Bundles and Perturbs Mineral Nutrient Assimilation in Rice. Sci Rep, 5: 18609.
Undan J. R., Tamiru M., Abe A., Yoshida K., Kosugi S., Takagi H., Yoshida K., Kanzaki H., Saitoh H., Fekih R., Sharma S., Undan J., Yano M., Terauchi R. 2012. Mutation in OsLMS, a gene encoding a protein with two double-stranded RNA binding motifs, causes lesion mimic phenotype and early senescence in rice Oryza sativa L.. Genes Genet Syst, 873: 169-179.
Vega-Sanchez M. E., Zeng L., Chen S., Leung H., Wang G. L. 2008. SPIN1, a K homology domain protein negatively regulated and ubiquitinated by the E3 ubiquitin ligase SPL11, is involved in flowering time control in rice. Plant Cell, 206: 1456-1469.
Wang J., Ye B., Yin J., Yuan C., Zhou X., Li W., He M., Wang J., Chen W., Qin P., Ma B., Wang Y., Li S., Chen X. 2015a. Characterization and fine mapping of a light-dependent leaf lesion mimic mutant 1 in rice. Plant Physiol Biochem, 97: 44-51.
Wang L., Pei Z., Tian Y., He C. 2005. OsLSD1, a rice zinc finger protein, regulates programmed cell death and callus differentiation. Mol Plant Microbe Interact, 185: 375-384.
Wang S. H., Lim J. H., Kim S. S., Cho S. H., Yoo S. C., Koh H. J., Sakuraba Y., Paek N. C. 2015b. Mutation of SPOTTED LEAF3 SPL3 impairs abscisic acid-responsive signalling and delays leaf senescence in rice. J Exp Bot, 6622: 7045-7059.
Wang S., Lei C., Wang J., Ma J., Tang S., Wang C., Zhao K., Tian P., Zhang H., Qi C., Cheng Z., Zhang X., Guo X., Liu L., Wu C., Wan J. 2017. SPL33, encoding an eEF1A-like protein, negatively regulates cell death and defense responses in rice. J Exp Bot, 685: 899-913.
Wang Z., Wang Y., Hong X., Hu D., Liu C., Yang J., Li Y., Huang Y., Feng Y., Gong H., Li Y., Fang G., Tang H., Li Y. 2015c. Functional inactivation of UDP-N-acetylglucosamine pyrophosphorylase 1 UAP1 induces early leaf senescence and defence responses in rice. J Exp Bot, 663: 973-987.
Xiao G., Zhou J., Lu X., Huang R., Zhang H. 2018. Excessive UDPG resulting from the mutation of UAP1 causes programmed cell death by triggering reactive oxygen species accumulation and caspase-like activity in rice. New Phytol, 2171: 332-343.
Yamanouchi U., Yano M., Lin H., Ashikari M., Yamada K. 2002. A rice spotted leaf gene, Spl7, encodes a heat stress transcription factor protein. Proc Natl Acad Sci U S A, 9911: 7530-7535.
You Q., Zhai K., Yang D., Yang W., Wu J., Liu J., Pan W., Wang J., Zhu X., Jian Y., Liu J., Zhang Y., Deng Y., Li Q., Lou Y., Xie Q., He Z. 2016. An E3 Ubiquitin Ligase-BAG Protein Module Controls Plant Innate Immunity and Broad-Spectrum Disease Resistance. Cell Host Microbe, 206: 758-769.
Yuan Y., Zhong S., Li Q., Zhu Z., Lou Y., Wang L., Wang J., Wang M., Li Q., Yang D., He Z. 2007. Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol J, 52: 313-324.
Zeng L. R., Qu S., Bordeos A., Yang C., Baraoidan M., Yan H., Xie Q., Nahm B. H., Leung H., Wang G. L. 2004. Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell, 1610: 2795-2808.
Zhang X. B., Feng B. H., Wang H. M., Xu X., Shi Y. F., He Y., Chen Z., Sathe A. P., Shi L., Wu J. L. 2018. A substitution mutation in OsPELOTA confers bacterial blight resistance by activating the salicylic acid pathway. J Integr Plant Biol, 602: 160-172.
Zhao J., Liu P., Li C., Wang Y., Guo L., Jiang G., Zhai W. 2017. LMM5.1 and LMM5.4, two eukaryotic translation elongation factor 1A-like gene family members, negatively affect cell death and disease resistance in rice. J Genet Genomics, 442: 107-118.
Zhou Q., Zhang Z., Liu T., Gao B., Xiong X. 2017. Identification and Map-Based Cloning of the Light-Induced Lesion Mimic Mutant 1 LIL1 Gene in Rice. Front Plant Sci, 8: 2122.
Zhu X., Yin J., Liang S., Liang R., Zhou X., Chen Z., Zhao W., Wang J., Li W., He M., Yuan C., Miyamoto K., Ma B., Wang J., Qin P., Chen W., Wang Y., Wang W., Wu X., Yamane H., Zhu L., Li S., Chen X. 2016. The Multivesicular Bodies MVBs-Localized AAA ATPase LRD6-6 Inhibits Immunity and Cell Death Likely through Regulating MVBs-Mediated Vesicular Trafficking in Rice. PLoS Genet, 129: e1006311.
(Managing Editor: Fang Hongmin)
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
|
| [34] |
|
| [35] |
|
| [36] |
|
| [37] |
|
| [38] |
|
| [39] |
|
| [40] |
|
| [41] |
|
| [42] |
|
| [43] |
|
| [44] |
|
| [45] |
|
| [46] |
|
| [47] |
|
| [48] |
|
| [49] |
|
| [50] |
|
| [51] |
|
| [52] |
|
| [53] |
|
| [54] |
|
| [55] |
|
| [56] |
|
| [57] |
|
| [58] |
|
| [59] |
|
| [60] |
|
| [61] |
|
| [62] |
|
| [63] |
|
| [64] |
|
| [65] |
|
| [66] |
|
| [67] |
|
| [68] |
|
| [69] |
|
| [70] |
|
| [71] |
|
| [72] |
|
| [73] |
|
| [74] |
|
| [75] |
|
| [76] |
|
| [77] |
|
| [78] |
|
| [79] |
|
| [80] |
|
| [81] |
|
| [82] |
|
| [83] |
|
| [84] |
|