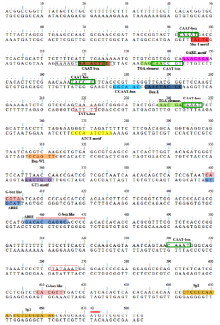
The promoter region of a drought and abscisic acid (ABA) inducible gene, osr40c1, was isolated from a salt-tolerant indica rice variety Pokkali, which is 670 bp upstream of the putative translation start codon. In silico promoter analysis of resulted sequence showed that at least 15 types of putative motifs were distributed within the sequence, including two types of common promoter elements, TATA and CAAT boxes. Additionally, several putative cis-acing regulatory elements which may be involved in regulation of osr40c1 expression under different conditions were found in the 5′-upstream region of osr40c1. These are ABA-responsive element, light-responsive elements (ATCT-motif, Box I, G-box, GT1-motif, Gap-box and Sp1), myeloblastosis oncogene response element (CCAAT-box), auxin responsive element (TGA-element), gibberellin-responsive element (GARE-motif) and fungal-elicitor responsive elements (Box E and Box-W1). A putative regulatory element, required for endosperm-specific pattern of gene expression designated as Skn-1 motif, was also detected in the Pokkali osr40c1promoter region. In conclusion, the bioinformatic analysis of osr40c1 promoter region isolated from indica rice variety Pokkali led to the identification of several important stress-responsive cis-acting regulatory elements, and therefore, the isolated promoter sequence could be employed in rice genetic transformation to mediate expression of abiotic stress induced genes.
The major cereal crops, rice, wheat and maize, provide more than half of the calories consumed by humans, worldwide (Langridge and Fleury, 2011). Rice (Oryza sativa L.), as a staple food for more than one-half of the world’ s population, provides approximately 23% of calorie needs (Khush, 2001). However, it has been predicted that the world may require 25% more rice production by the year 2030 to feed the growing population (Wani and Sah, 2014). To meet this challenge, we need to extend rice growing areas or develop improved rice varieties through breeding efforts to increase yield per hectare. The latter seems to be the only possible way to boost rice production as the availability of land for agriculture is becoming a limiting factor due to increasing population and urbanization (Ansari et al, 2015).
Among the factors that cause marked reduction in agriculture production globally, abiotic and biotic stress conditions have received a greater attention. A total of 50%-70% reduction in crop productivity and growth worldwide is reported due to abiotic stress factors, predominately salinity, drought, cold and submergence (Mittler, 2006). Therefore, there is a need for development of improved crop varieties that can withstand adverse environmental conditions to ensure the security of global food production.
In recent decades, considerable amount of effort has been directed towards developing transgenic rice lines that can withstand high levels of drought and salt stress conditions through constitutive over-expression of stress-responsive genes. Although this seems to be a promising approach to develop improved varieties resistant to abiotic stress, constitutive over- expression of genes including transcription factors (TFs) has been reported to cause undesirable side-effects such as stunted growth and reduced yield in transgenic tomato, tobacco and Arabidopsis plants (Kasuga et al, 1999, 2004; Hsieh et al, 2002; Youm et al, 2008). Therefore, the selection and use of suitable promoters is vital for regulation of introduced transgenes. This would minimize undesirable side-effects that may result due to constitutive promoter driven over-expression of genes in transformed lines (Kasuga et al, 2004). Furthermore, identification of cis-acting regulatory elements present in promoter sequences is essential as information from such studies would provide valuable insight into the underlying mechanism of promoter activity and transcriptional regulation of introduced gene(s) (Huda et al, 2013). Several categories of plant promoters have been described, including constitutive promoters, tissue/stage- specific promoters (spatiotemporal promoters), inducible promoters and synthetic promoters (Hernandez-Garcia and Finer, 2014), of which constitutive promoters allow transgene to express in most of the plant tissues at any developmental stage. These types of promoter sequences are generally derived from either plant viruses i.e. cauliflower mosaic virus (CaMV) 35S or house-keeping genes i.e. maize ubiquitin promoters, and are widely used in development of transgenic plants (Hernandez-Garcia and Finer, 2014; Shah et al, 2015). In contrast, inducible/controlled gene expression systems are used when a more precise regulation of transgene expression is required. These systems have been reported in many plant species including tobacco, rice and Arabidopsis (Borghi, 2010). In such systems, tissue-specific and inducible promoters are highly desirable for defined spatial and temporal expression of transgene in transgenic plants. For instance, tissue- or developmental stage-specific pattern of transgene expression is achieved by the use of tissue-specific promoters. Numerous tissue-specific promoters have been discovered in plants, such as root-specific (Chen et al, 2015), seed-specific (Furtado and Henry, 2005; Pan et al, 2015), and anther-specific promoters (Jeon et al, 1999). Inducible gene promoters are key regulators that would permit regulation of transgene expression in response to external chemical or physical stimuli, i.e. biotic and abiotic stress, plant hormones and pathogens (Hernandez-Garcia and Finer, 2014; Shah et al, 2015). Compared to other three native promoter types [constitutive, tissue/stage-specific (spatiotemporal) and inducible promoters], synthetic promoters are unique and can synthesize artificially to drive diverse transgene expression patterns (Hernandez-Garcia and Finer, 2014).
Although the CaMV 35S promoter allows transgene(s) to express at all the time anywhere in the plant, it can alter the expression pattern of the transgene(s). Moreover, there is a risk of releasing these constitutive promoters into the environment via horizontal gene transfer (Shah et al, 2015). Therefore, inducible gene expression systems have shown to be more beneficial in plant genetic engineering overcoming detrimental effects of constitutive over-expression of transgene(s). In most of rice transgenic studies, the transgene constructs are placed under the control of constitutive promoters, particularly CaMV 35S (Sundheep et al, 2008). Therefore, use of abiotic stress inducible promoters, which are activated only at abiotic stress conditions, provides an ideal approach in development of abiotic stress tolerant plants. This would allow us to over-come the potential drawbacks associated with utilization of constitutive promoters.
As a signaling molecule, plant phytohormone abscisic acid (ABA) plays a vital role in mediating stress regulatory network particularly in response to drought and salinity stress. A large number of TFs, involved in abiotic stress signaling network in plants, have been described. Examples of common TF families, involved in abiotic stress signaling, are ABA-responsive element binding proteins (ABRE), myelocytomatosis oncogene (MYC), myeloblastosis oncogene (MYB), cold-binding factor/dehydration-responsive element binding (CBF/DREB), apetala2 (AP2)/ethylene-responsive element binding protein (EREBP), NAC and zinc-finger homeodomain (ZF-HD) (Lata et al, 2011). Of these, members of the ABRE, MYC and MYB TF families are mainly involved in ABA-dependent stress signaling pathways. Furthermore, the cis-acting regulatory elements or the TF binding sites (TFBs) distributed in upstream gene sequences are also critical for the regulation of gene(s) at the transcriptional level. These elements interact with TFs for fine regulation of gene expression. Therefore, identification and functional validation of these cis-acting regulatory elements present in particularly stress-responsive gene promoter sequences would provide insight into their transcriptional regulation under stress conditions. Several stress-responsive elements have been well characterized in plants, including dehydration responsive element (DRE, A/GCCGAC) (Yamaguchi-Shinozaki and Shinozaki, 1994), low temperature responsive C-repeat binding factor (CBF) (Jiang et al, 1996) and ABRE (PyACGTGGC) (Yamaguchi-Shinozaki and Shinozaki, 2006). For instance, ABA-inducible basic leucine zipper (bZIP) TFs, ABRE-binding proteins (AREB) i.e. AREB1 and AREB2, bind to conserved ABRE motifs are present in promoter sequence of Arabidopsis dehydration-responsive RD29B gene and involved in activation of RD29B in response to ABA (Uno et al, 2000).
The subsets of genes that are differentially expressed under abiotic stress conditions provide a valuable source for isolation and functional characterization of stress inducible promoters and their regulatory elements (Hernandez-Garcia and Finer, 2014). These promoters would be served as ideal candidates for transcriptional regulation of transgenes in the development of abiotic stress tolerant transgenic lines. The present study describes isolation and sequence analysis of stress-responsive osr40c1 gene promoter from leaf tissues of Pokkali (O. sativa ssp. indica), which is a salinity tolerant rice variety. The osr40c1 (1 458 bp) encodes an ABA-responsive protein (40 kDa) (Moons et al, 1997) that belongs to OSR40C protein family. There are three members in this protein family, namely OSR40C1, OSR40G2 and OSR40G3 (Moons et al, 1997), among which, osr40c1 transcripts shows a root-specific expression in response to ABA and salt stress, while osr40g2 and osr40g3 show shoot-specific (Moons et al, 1997). The isolated osr40c1 promoter sequence from Pokkali together with the information in this report will be useful in development of rice transgenic lines against abiotic stress.
Seeds of indica rice variety Pokkali (salinity resistant) were obtained from the Rice Research and Development Institute, Bathalagoda, Sri Lanka, and grown in a greenhouse (temperature of 27 º C-29 º C and relative humidity of 70%-80%) in the University of Colombo, Sri Lanka.
Genomic DNA was extracted from leaves of Pokkali using the cetyl trimethyl ammonium bromide (CTAB) method described by Doyle and Doyle (1987). In brief, 0.8 g leaf tissues washed with 70% ethanol were ground into a fine powder using liquid nitrogen, then transferred into an Eppendorf tube containing pre-heated (65 º C) 2× CTAB buffer (700 µ L) and centrifuged at 8 000 r/min for 5 min. Subsequently, 5 µ L RNase A (10 mg/mL) was added to the resultant supernatant and incubated at 56 º C for 30 min. An equal volume of chloroform : isopropyl alcohol (24 : 1) was added and swirled at the end of incubation period. The mixture was then centrifuged at 12 000 r/min for 2 min, and 500 µ L isopropanol was added to the aqueous phase. The content was incubated at -20 º C for 30 min which was then centrifuged at 10 000 r/min for 5 min. Thereafter, the nucleic acid containing pellet was washed several times with 70% ethanol (500 µ L). The drained and dried pellet was re-suspended in 60 µ L of sterilized distilled water.
PCR reactions were performed in a 50 µ L reaction volume containing 2 µ L template DNA, 5 µ L of each forward and reverse primers, 1 µ L of 10 mmol/L dNTPs, 0.5 µ L of Taq DNA polymerase, 5 µ L of 10× buffer containing MgCl2 (Promega) and 31.5 µ L water in a BIORAD MyCycler® . To amplify osr40c1 promoter region, following primer sequences were designed, introducing a PstI restriction site (CTGCAG) and a BamHI restriction site (GTCGAC) in the forward (F1, ATTCTGCAGACGGCCGGTTTA) and reverse (R1, ATTGTC GACCGAAGCCGAACA) primers (sites are underlined), respectively. These primers were designed and synthesised on the basis of the published O. sativa japonica group (Nipponbare) chromosome 3 clone OSJNBb0014I10, complete sequence (GenBank accession number: AC126222.2). The Primer3 version 0.4.0 tool (http://bioinfo.ut.ee/primer3-0.4.0/) was used to design primer sequences on the basis of putative Nipponbare osr40c1 sequence.
The following parameters were used during the PCR amplification: 94 º C for 4 min; 35 cycles: 94 º C for 1 min, 56 º C for 1 min and 72 º C for 1 min; final extension: 72 º C for 7 min. At the end of the PCR amplification, aliquots of amplified products were loaded onto a 2% agarose gel and subjected to gel electrophoresis.
The amplified PCR products were purified using the PureLink® PCR Purification Kit (Thermo Fisher Scientific, Massachusetts, USA). In brief, amplified products were excised from the agarose gel under the UV trans-illuminator and transferred into an Eppendorf tube containing 200 µ L capture buffer. Contents were then incubated at 65 º C for 15 min and centrifuged at 13 000 r/min for 1 min in a spin column. Resulted pellet was centrifuged again with 700 µ L wash buffer at 13 000 r/min for twice. Subsequently, the column was dried by centrifuging at 13 000 r/min for 1 min. Pre-heated deionized water was added to the column and incubated for another 3 min at 65 º C. At the end of incubation, content was centrifuged at 13 000 r/min for 1 min.
Purified PCR product was dried in a vacuum centrifuge, and re-dissolved in deionized water (12 µ L). Restriction enzymes (PstI and BamHI restriction enzymes) digested PCR product was ligated to PstI and BamHI cleaved pUC18 and then transformed into Escherichia coli strain GM2163. The ligation reaction was carried out in a 20 µ L volume containing 8 µ L digested pUC18 plasmid DNA, 4 µ L digested PCR product, 2 µ L ligase buffer, 0.25 µ L ligase enzyme and 5.75 µ L deionized water. The reaction mixture was incubated at 10 º C overnight. Bacterial transformation was performed using the heat shock method described by Hanahan (1983), and positive transformants were selected through colony PCR. Recombinant pUC18 plasmid containing osr40c1 promoter region was isolated using the alkaline lysis method for sequencing.
To obtain the full-length nucleotide sequence amplified with F1 and R1 primers, pUC18 backbone based primers (pUC18_F1, GTAAAACGACGGCCAGT and pUC18_R1, CAGGAAACAGCTATGGC) were used. The DNA sequencing was performed using an automate sequencer (ABI310 DNA sequencer) at a commercial sequencing service.
By using the previously published osr40c1 mRNA sequence as the query sequence (GenBank accession number: X95402), putative O. sativa ssp. indica osr40c1 with a gene identifier, OSINDICA_03G20090, was identified. The upstream genomic region (1 kb) of this gene was extracted from PLAZA 3.0 monocot platform (http://bioinformatics.psb.ugent.be/plaza/) and aligned with the sequenced Pokkali osr40c1 promoter region, using the multiple sequence alignment tool in the CLC Sequence Viewer 6 (CLC Bio).
In addition, putative osr40g2 and osr40g3 sequences of published indica and japonica genomes were identified using the previously published O. sativa osr40g2 (GenBank accession: Y08987) and osr40g3 (GenBank accession: Y08988) transcript sequences as queries using the BLAST (Basic Local Alignment Search Tool) search tool in the PLAZA 3.0 monocot platform. The genomic and coding sequences of osr40c family were extracted from monocot PLAZA 3.0 tool and gene structures were visualized using the Gene Structure Display Server 2.0 (GSDS 2.0; http://gsds.cbi.pku.edu.cn/). Gene expression data for osr40c1 in response to stress conditions were obtained from the Genevestigator database (https://genevestigator.com/ gv/). Furthermore, the sequenced osr40c1 promoter region (670 bp) was analysed for the presence of potential cis-acting regulatory elements using the PlantCARE database (Lescot et al, 2002) (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
The chromosomal locations of osr40cfamily members reported by Moons et al (1997) were analysed using the published transcript sequences as query sequences. Both in indica and japonica genomes, osr40c1 was located on chromosome 3 (negative strand) with gene identifiers, OSINDICA_03G20090 and OS03G21040, respectively (Table 1). Furthermore, both osr40g2 and osr40g3 were located on chromosome 7 (positive strand) (Table 1). Gene structures of putative osr40c family members identified in O. sativa ssp. indica and the ortholog(s) sequences identified in O. sativa ssp. japonica are shown in Fig. 1.
| Table 1 Descriptions of osr40c gene family members in rice. |
In silico expression analysis of osr40c1 in rice plants using the Genevestigator database revealed enhanced expression of this gene in response to multiple stress responses (Table 2). However, the promoter region of this stress-responsive gene has not been studied thus far.
| Table 2 Expression of osr40c1 in rice plants in response to stress conditions from the Genevestigator database. |
In the present study, approximately a 683 bp long fragment was produced from the PCR amplification of Pokkali genomic DNA with F1 and R1 primers. The sequence of this amplified Pokkali osr40c1 promoter region was aligned with the 1 kb upstream genomic sequence of OSINDICA_03G20090 gene (putative indica osr40c1) and as expected this showed 100% sequence similarity. The sequence of the isolated Pokkali osr40c1 promoter region has been deposited at the GenBank database under the accession number KP203842.1.
Prediction of cis-acting regulatory elements distributed in isolated Pokkali osr40c1 promoter was performed using the PlantCARE database (Lescot et al, 2002). It showed the presence of at least 15 types of known cis-acting regulatory elements in the isolated osr40c1 promoter. The promoter sequence along with positions of detected putative cis-acting regulatory elements is presented in Fig. 2. The description and abundance of those elements are listed in Table 3.
| Table 3 Description of putative cis-acting regulatory elements in Pokkali osr40c1 promoter region from the PlantCARE database. |
The OSR40C family was first described by Moons et al (1997). However, this gene family has not received much attention. The availability of genome sequence of many plant species including the model monocot, O. sativa ssp. indica (Yu et al, 2002) and O. sativa ssp. japonica (International Rice Genome Sequencing Project, 2005; Kawahara et al, 2013), and easy accessibility to a number of web-based genome browsers and databases have opened-up ways to investigate this gene family in detail.
As observed in many stress-responsive gene families (Lan et al, 2013), low intron numbers were observed in all putative members of the osr40c family identified in indica and japonica rice. For instance, putative osr40c1 in both indica and japonica rice had only two introns (Fig. 1-A). Furthermore, it was found that its expression is induced by multiple stress conditions.
TFBs or cis-acting regulatory elements located in the genomic region upstream of a 5′ -untraslated region of a gene provide binding sites for specific TFs. Thus, investigation of cis-acting regulatory elements present in promoter sequences provides insight into fine regulation of gene expression under different conditions. In silico promoter analysis of the Pokkali osr40c1 promoter sequence identified core elements such as TATA-box and CAAT-box. In addition, numerous cis-acting regulatory elements that allow inducible pattern of osr40c1 expression were also detected in the promoter region, including ABREs, light-responsive motifs (ATCT-motif, Box I, G-box like motif, GT1-motif, Gap-box and Sp1), hormone-responsive elements (TGA-element and GARE-motif), fungal inducible elements (Box E and Box-W1) and MYB response element (CCAAT-box). Additionally, a cis-acting regulatory element found in genes that display an endosperm specific pattern of expression (Skn-1 motif) was also detected in the Pokkali-osr40c1 promoter sequence. It has been reported that ABREs are important in ABA-mediated signal transduction of several dehydration-responsive genes in Arabidopsis i.e. RD29B (Uno et al, 2000).
Pokkali osr40c1 promoter region also contains three putative G-box like elements that closely resemble ACGT-containing ABREs. These G-box like motifs serve as binding sites for specific bZIP proteins known as G-box binding proteins and have been found in many gene promoters that respond to environmental stimuli such as UV light, ABA, anaerobiosis, red light and wounding (Nakagawa et al, 1996; Uno et al, 2000). For instance, it has shown that G-box like motifs are needed for the expression of wheat Em gene in response to ABA (Nantel and Quatrano, 1996).
Some of the predicted cis-acting regulatory elements in the Pokkali osr40c1 promoter region have previously been reported in promoter regions of osr40g2 and osr40g3 isolated from Nona Bokra (O. sativa ssp. indica) (Moons et al, 1997). For instance, MYB response element and G-box like elements were commonly found in Pokkali osr40c1 and Nona Bokra osr40g2 promoter regions, and ACGT-containing ABREs were present in both Pokkali osr40c1 and Nona Bokra osr40g3 promoters.
The present study successfully isolated and sequenced the putative osr40c1 promoter region from a salt tolerant rice variety Pokkali. Moreover, in silico promoter analysis revealed presence of numerous stress-responsive cis-acting regulatory elements in the isolated promoter sequence. The findings reported in this study together with the evidences of enhanced transcription of osr40c1 in rice plants in response to multiple stress conditions would provide insights into the stress inducible nature of the osr40c1 promoter, and therefore, could be served as a valuable source for stress tolerant transgenic rice breeding in the future. However, follow-up studies are needed to functionally validate the isolated osr40c1 promoter activity through transgenic methods and expression studies.
The authors have declared that no competing interests exist.
| 1 |
|
| 2 |
|
| 3 |
|
| 4 |
|
| 5 |
|
| 6 |
|
| 7 |
|
| 8 |
|
| 9 |
|
| 10 |
|
| 11 |
|
| 12 |
|
| 13 |
|
| 14 |
|
| 15 |
|
| 16 |
|
| 17 |
|
| 18 |
|
| 19 |
|
| 20 |
|
| 21 |
|
| 22 |
|
| 23 |
|
| 24 |
|
| 25 |
|
| 26 |
|
| 27 |
|
| 28 |
|
| 29 |
|
| 30 |
|
| 31 |
|
| 32 |
|
| 33 |
|