
Rice Science ›› 2023, Vol. 30 ›› Issue (6): 652-660.DOI: 10.1016/j.rsci.2023.06.005
• Research Papers • Previous Articles
Lin Shaodan1,2, Yao Yue1,3, Li Jiayi1,3, Li Xiaobin1,3, Ma Jie1,3, Weng Haiyong1,3, Cheng Zuxin1,4( ), Ye Dapeng1,3(
), Ye Dapeng1,3( )
)
Received:2023-02-28
Accepted:2023-06-30
Online:2023-11-28
Published:2023-08-10
Contact:
YE Dapeng (ydp@fafu.edu.cn);
CHENG Zuxin (chengzuxin@163.com)
Lin Shaodan, Yao Yue, Li Jiayi, Li Xiaobin, Ma Jie, Weng Haiyong, Cheng Zuxin, Ye Dapeng. Application of UAV-Based Imaging and Deep Learning in Assessment of Rice Blast Resistance[J]. Rice Science, 2023, 30(6): 652-660.
Add to citation manager EndNote|Ris|BibTeX
| Backbone | Training time (h) | Box loss | Mask loss | Classification loss | Segmentation loss |
|---|---|---|---|---|---|
| ResNet50 | 60.88 | 0.024 | 0.024 | 0.069 | 0.022 |
| ResNet101 | 36.22 | 0.026 | 0.026 | 0.083 | 0.022 |
| Swin-Tiny | 30.23 | 0.097 | 0.065 | 0.040 | 0.044 |
| Swin-Base | 38.27 | 0.067 | 0.044 | 0.027 | 0.034 |
Table 1. RiceblastSegMask training parameters using different backbone networks.
| Backbone | Training time (h) | Box loss | Mask loss | Classification loss | Segmentation loss |
|---|---|---|---|---|---|
| ResNet50 | 60.88 | 0.024 | 0.024 | 0.069 | 0.022 |
| ResNet101 | 36.22 | 0.026 | 0.026 | 0.083 | 0.022 |
| Swin-Tiny | 30.23 | 0.097 | 0.065 | 0.040 | 0.044 |
| Swin-Base | 38.27 | 0.067 | 0.044 | 0.027 | 0.034 |
| Disease level | ResNet50 | ResNet101 | Swin-Tiny | Swin-Base | ResNet50 | ResNet101 | Swin-Tiny | Swin-Base |
|---|---|---|---|---|---|---|---|---|
| Box average precision | Mask average precision | |||||||
| 1 | 100.00 a | 100.00 a | 80.00 | 100.00 a | 90.00 | 97.33 a | 60.00 | 80.00 |
| 2 | 98.88 a | 98.86 | 98.01 | 98.77 | 95.95 a | 95.42 | 90.52 | 93.07 |
| 3 | 97.88 | 98.16 | 98.21 a | 97.97 | 94.65 | 95.46 a | 91.13 | 93.96 |
| 4 | 98.99 | 99.00 a | 98.29 | 98.98 | 97.27 a | 96.73 | 91.45 | 93.98 |
| 5 | 100.00 a | 100.00 a | 100.00 a | 100.00 a | 100.00 a | 95.05 | 95.05 | 95.05 |
| 6 | 98.99 | 99.00 a | 98.42 | 99.00 a | 97.04 a | 96.80 | 91.47 | 93.91 |
| 7 | 98.80 a | 98.77 | 97.90 | 98.74 | 96.26 a | 95.59 | 90.17 | 92.95 |
| 8 | 97.97 a | 97.85 | 97.29 | 97.86 | 93.99 a | 93.71 | 89.31 | 91.08 |
| 9 | 98.87 | 98.89 | 98.25 | 98.98 a | 97.55 | 97.33 | 92.76 | 97.56 a |
Table 2. Box average precision and mask average precision of instance segmentation at different rice blast levels with RiceblastSegMask model using different backbone networks.
| Disease level | ResNet50 | ResNet101 | Swin-Tiny | Swin-Base | ResNet50 | ResNet101 | Swin-Tiny | Swin-Base |
|---|---|---|---|---|---|---|---|---|
| Box average precision | Mask average precision | |||||||
| 1 | 100.00 a | 100.00 a | 80.00 | 100.00 a | 90.00 | 97.33 a | 60.00 | 80.00 |
| 2 | 98.88 a | 98.86 | 98.01 | 98.77 | 95.95 a | 95.42 | 90.52 | 93.07 |
| 3 | 97.88 | 98.16 | 98.21 a | 97.97 | 94.65 | 95.46 a | 91.13 | 93.96 |
| 4 | 98.99 | 99.00 a | 98.29 | 98.98 | 97.27 a | 96.73 | 91.45 | 93.98 |
| 5 | 100.00 a | 100.00 a | 100.00 a | 100.00 a | 100.00 a | 95.05 | 95.05 | 95.05 |
| 6 | 98.99 | 99.00 a | 98.42 | 99.00 a | 97.04 a | 96.80 | 91.47 | 93.91 |
| 7 | 98.80 a | 98.77 | 97.90 | 98.74 | 96.26 a | 95.59 | 90.17 | 92.95 |
| 8 | 97.97 a | 97.85 | 97.29 | 97.86 | 93.99 a | 93.71 | 89.31 | 91.08 |
| 9 | 98.87 | 98.89 | 98.25 | 98.98 a | 97.55 | 97.33 | 92.76 | 97.56 a |
| Model | AP (IOU 0.50) | AP (IOU 0.75) | AP (IOU 0.90) | mAP (IOU 0.50‒0.95) |
|---|---|---|---|---|
| YOLACT | 92.09 | 90.50 | 48.78 | 77.12 |
| YOLACT++ | 93.10 | 90.53 | 49.86 | 77.83 |
| Fast R-CNN | 87.02 | 82.03 | 33.56 | 67.54 |
| Mask R-CNN | 95.03 | 87.14 | 57.35 | 79.84 |
| Transfiner | 95.08 | 85.33 | 58.33 | 79.58 |
| RiceblastSegMask | 96.83 | 87.33 | 56.33 | 80.16 |
Table 3. Average precision of different target detection models. %
| Model | AP (IOU 0.50) | AP (IOU 0.75) | AP (IOU 0.90) | mAP (IOU 0.50‒0.95) |
|---|---|---|---|---|
| YOLACT | 92.09 | 90.50 | 48.78 | 77.12 |
| YOLACT++ | 93.10 | 90.53 | 49.86 | 77.83 |
| Fast R-CNN | 87.02 | 82.03 | 33.56 | 67.54 |
| Mask R-CNN | 95.03 | 87.14 | 57.35 | 79.84 |
| Transfiner | 95.08 | 85.33 | 58.33 | 79.58 |
| RiceblastSegMask | 96.83 | 87.33 | 56.33 | 80.16 |

Fig. 3. Representative unmanned aerial vehicle (UAV)-based images for rice blast resistance grading among different genotypes using RiceblastSegMask. The numbers in the figures represent the disease levels of rice blast.
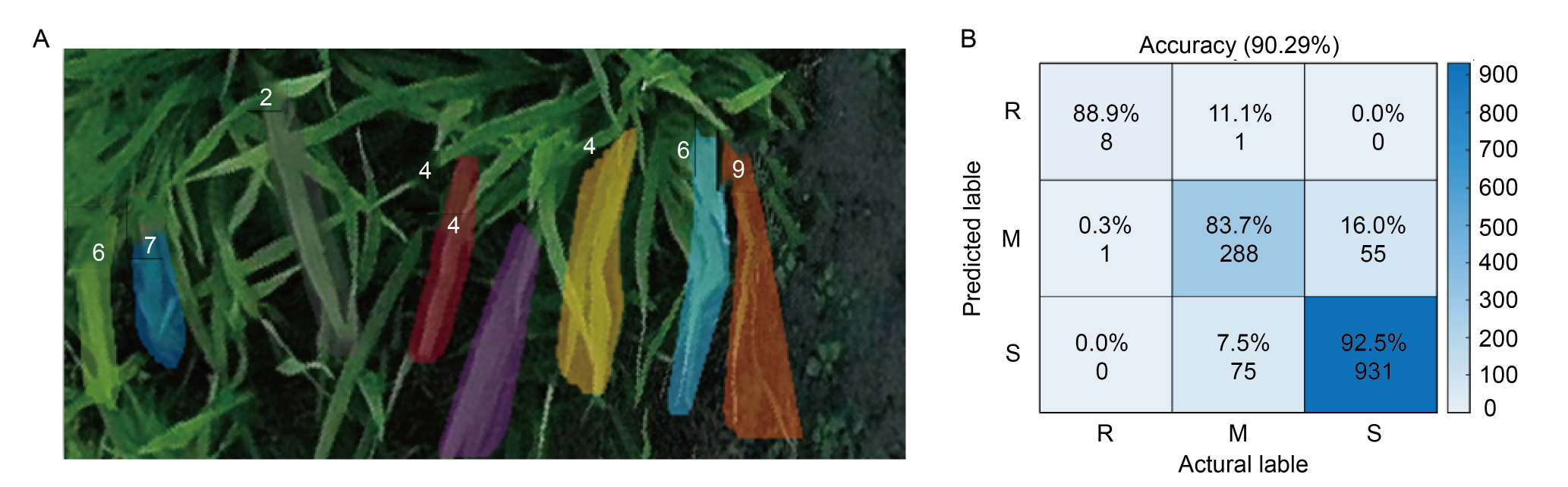
Fig. 4. Instance segmentation of rice blast based on RiceblastSegMask (A) and confusion matrix of rice blast resistance assessment (B). The numbers in the figures represent the disease levels of rice blast. R, Resistant; M, Moderately resistant; S, Susceptible.
| [1] | Alvarez J, Martinez E, Diezma B. 2021. Application of hyperspectral imaging in the assessment of drought and salt stress in magneto-primed Triticale seeds. Plants, 10(5): 835. |
| [2] | An G Q, Xing M F, He B B, Kang H Q, Shang J L, Liao C H, Huang X D, Zhang H G. 2021. Extraction of areas of rice false smut infection using UAV hyperspectral data. Remote Sens, 13(16): 3185. |
| [3] | Anderson R S. 1991. The origins of the International Rice Research Institute. Minerva, 29(1): 61-89. |
| [4] | Araus J L, Cairns J E. 2014. Field high-throughput phenotyping: The new crop breeding frontier. Trends Plant Sci, 19(1): 52-61. |
| [5] | Ariza-Sentís M, Valente J, Kooistra L, Kramer H, Mücher S. 2023. Estimation of spinach (Spinacia oleracea) seed yield with 2D UAV data and deep learning. Smart Agric Technol, 3: 100129. |
| [6] | Atkins J. 1967. An international set of rice varieties for differentiating races of Piricularia oryzae. Phytopathology, 57: 297-301. |
| [7] | Bal S S, Kumar S, Kar A, Mishra I, Panda P. 2019. Screening of mungbean germplasm for resistance against important diseases under rice fallow condition. J Plant Prot Environ, 16(1): 34-37. |
| [8] | Bolya D, Zhou C, Xiao F Y, Lee Y J. 2020. YOLACT: Real-time instance segmentation. 2019 IEEE/CVF International Conference on Computer Vision (ICCV). October 27-November 2, 2019. Seoul, Korea (South): IEEE: 9156-9165. |
| [9] | Brugger A, Schramowski P, Paulus S, Steiner U, Kersting K, Mahlein A K. 2021. Spectral signatures in the UV range can be combined with secondary plant metabolites by deep learning to characterize barley-powdery mildew interaction. Plant Pathol, 70(7): 1572-1582. |
| [10] | Cai N, Zhou X G, Yang Y B, Wang J, Zhang D Y, Hu R J. 2018. Use of UAV images to assess narrow brown leaf spot severity in rice. Int J Precis Agric Aviat, 1(1): 38-42. |
| [11] | Cen H Y, Wan L, Zhu J P, Li Y J, Li X R, Zhu Y M, Weng H Y, Wu W K, Yin W X, Xu C, Bao Y D, Feng L, Shou J Y, He Y. 2019. Dynamic monitoring of biomass of rice under different nitrogen treatments using a lightweight UAV with dual image-frame snapshot cameras. Plant Methods, 15(1): 1-16. |
| [12] | Feng S, Zhao D X, Guan Q, Li J P, Liu Z Y, Jin Z Y, Li G M, Xu T Y. 2022. A deep convolutional neural network-based wavelength selection method for spectral characteristics of rice blast disease. Comput Electron Agric, 199: 107199. |
| [13] | Feng X P, Zhan Y H, Wang Q, Yang X F, Yu C L, Wang H Y, Tang Z Y, Jiang D A, Peng C, He Y. 2020. Hyperspectral imaging combined with machine learning as a tool to obtain high- throughput plant salt-stress phenotyping. Plant J, 101(6): 1448-1461. |
| [14] | Jin P F, Wang Y, Tan Z, Liu W B, Miao W G. 2020. Antibacterial activity and rice-induced resistance, mediated by C15surfactin A, in controlling rice disease caused by Xanthomonas oryzae pv. oryzae. Pestic Biochem Physiol, 169: 104669. |
| [15] | Ke L, Danelljan M, Li X, Tai Y W, Tang C K, Yu F. 2022. Mask transfiner for high-quality instance segmentation. 2022 IEEE/ CVF Conference on Computer Vision and Pattern Recognition (CVPR). June 18-24, 2022. New Orleans, LA, USA: IEEE: 4402-4411. |
| [16] | Kim Y, Roh J H, Kim H. 2017. Early forecasting of rice blast disease using long short-term memory recurrent neural networks. Sustainability, 10(2): 34. |
| [17] | Laha G S, Singh R, Ladhalakshmi D, Sunder S, Prasad M S, Dagar C S, Babu V R. 2017. Importance and management of rice diseases: A global perspective. In: Chauhan B S, Jabran K, Mahajan G. Rice Production Worldwide. Cham, Switzerland: Springer International Publishing: 303-360. |
| [18] | Li D L, Bai D, Tian Y, Li Y H, Zhao C S, Wang Q, Guo S Y, Gu Y Z, Luan X Y, Wang R Z, Yang J L, Hawkesford M J, Schnable J C, Jin X L, Qiu L J. 2023. Time series canopy phenotyping enables the identification of genetic variants controlling dynamic phenotypes in soybean. J Integr Plant Biol, 65(1): 117-132. |
| [19] | Li W, Feng X S, Zha K W, Li S N, Zhu H S. 2021. Summary of target detection algorithms. J Phys: Conf Ser, 1757(1): 012003. |
| [20] | Li Y, Qian M Y, Liu P F, Cai Q, Li X Y, Guo J W, Yan H, Yu F Y, Yuan K, Yu J, Qin L M, Liu H X, Wu W, Xiao P Y, Zhou Z W. 2019. The recognition of rice images by UAV based on capsule network. Cluster Comput, 22(4): 9515-9524. |
| [21] | Liang J Y, Cao J Z, Sun G L, Zhang K, van Gool L, Timofte R. 2021. SwinIR: Image restoration using Swin Transformer. Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV) Workshops. 11-17 October, 2021. Montreal, BC, Canada: IEEE: 1833-1844. |
| [22] | Lin S D, Li X B, Yang B Y, Chen C, He W C, Weng H Y, Ye D P. 2022. Detecting citrus Huanglongbing from few-shot microscopic images using an improved DETR. Trans Chin Soc Agric Eng, 38(12): 216-223. (in Chinese with English abstract) |
| [23] | Ma X Q, Duan G H, Chen H F, Tang P, Su S Y, Wei Z X, Yang J. 2022. Characterization of infected process and primary mechanism in rice Acuce defense against rice blast fungus, Magnaporthe oryzae. Plant Mol Biol, 110(3): 219-234. |
| [24] | Nugent P W, Shaw J A, Jha P, Scherrer B, Donelick A, Kumar V. 2018. Discrimination of herbicide-resistant kochia with hyper- spectral imaging. J Appl Remote Sens, 12(1): 016037. |
| [25] | Oerke E C, Leucker M, Steiner U. 2019. Sensory assessment of Cercospora beticola sporulation for phenotyping the partial disease resistance of sugar beet genotypes. Plant Methods, 15(1): 1-12. |
| [26] | Shahriar S A, All Imtiaz A, Hossain M B, Husna A, Eaty M N K. 2020. Review: Rice blast disease. Annu Res Rev Biol, 35(1): 50-64. |
| [27] | Wan L, Li Y J, Cen H Y, Zhu J P, Yin W X, Wu W K, Zhu H Y, Sun D W, Zhou W J, He Y. 2018. Combining UAV-based vegetation indices and image classification to estimate flower number in oilseed rape. Remote Sens, 10(9): 1484. |
| [28] | Zhang D Y, Zhou X G, Zhang J, Lan Y B, Xu C, Liang D. 2018. Detection of rice sheath blight using an unmanned aerial system with high-resolution color and multispectral imaging. PLoS One, 13(5): e0187470. |
| [29] | Zhang J N, Feng X P, Wu Q G, Yang G F, Tao M Z, Yang Y, He Y. 2022. Rice bacterial blight resistant cultivar selection based on visible/near-infrared spectrum and deep learning. Plant Methods, 18(1): 1-16. |
| [30] | Zhou X, Zheng H B, Xu X Q, He J Y, Ge X K, Yao X, Cheng T, Zhu Y, Cao W X, Tian Y C. 2017. Predicting grain yield in rice using multi-temporal vegetation indices from UAV-based multi- spectral and digital imagery. ISPRS J Photogramm Remote Sens, 130: 246-255. |
| [1] | Zhou Longfei, Meng Ran, Yu Xing, Liao Yigui, Huang Zehua, Lü Zhengang, Xu Binyuan, Yang Guodong, Peng Shaobing, Xu Le. Improved Yield Prediction of Ratoon Rice Using Unmanned Aerial Vehicle-Based Multi-Temporal Feature Method [J]. Rice Science, 2023, 30(3): 247-256. |
| [2] | Wu Zhongling, Qiu Jiehua, Shi Huanbin, Lin Chuyu, Yue Jiangnan, Liu Zhiquan, Xie Wei, Naweed I. Naqvi, Kou Yanjun, Tao Zeng. Polycomb Repressive Complex 2-Mediated H3K27 Trimethylation Is Required for Pathogenicity in Magnaporthe oryzae [J]. Rice Science, 2022, 29(4): 363-374. |
| [3] | Suhas Gorakh Karkute, Vishesh Kumar, Mohd Tasleem, Dwijesh Chandra Mishra, Krishna Kumar Chaturvedi, Anil Rai, Amitha Mithra Sevanthi, Kishor Gaikwad, Tilak Raj Sharma, Amolkumar U. Solanke. Genome-Wide Analysis of von Willebrand Factor A Gene Family in Rice for Its Role in Imparting Biotic Stress Resistance with Emphasis on Rice Blast Disease [J]. Rice Science, 2022, 29(4): 375-384. |
| [4] | Zhou Ying, Wan Tao, Yuan Bin, Lei Fang, Chen Meijuan, Wang Qiong, Huang Ping, Kou Shuyan, Qiu Wenxiu, Liu Li. Improving Rice Blast Resistance by Mining Broad-Spectrum Resistance Genes at Pik Locus [J]. Rice Science, 2022, 29(2): 133-142. |
| [5] | Junhua Lu, Xuemei Yang, Jinfeng Chen, Tingting Li, Zijin Hu, Ying Xie, Jinlu Li, Jiqun Zhao, Mei Pu, Hui Feng, Jing Fan, Yanyan Huang, Jiwei Zhang, Wenming Wang, Yan Li. Osa-miR439 Negatively Regulates Rice Immunity Against Magnaporthe oryzae [J]. Rice Science, 2021, 28(2): 156-165. |
| [6] | Yanchang Luo, Tingchen Ma, Teo Joanne, Zhixiang Luo, Zefu Li, Jianbo Yang, Zhongchao Yin. Marker-Assisted Breeding of Thermo-Sensitive Genic Male Sterile Line 1892S for Disease Resistance and Submergence Tolerance [J]. Rice Science, 2021, 28(1): 89-98. |
| [7] | Ning Xiao, Yunyu Wu, Aihong Li. Strategy for Use of Rice Blast Resistance Genes in Rice Molecular Breeding [J]. Rice Science, 2020, 27(4): 263-277. |
| [8] | B. ANGELES-SHIM Rosalyn, P. REYES Vincent, M. del VALLE Marilyn, S. LAPIS Ruby, SHIM Junghyun, SUNOHARA Hidehiko, K. JENA Kshirod, ASHIKARI Motoyuki, DOI Kazuyuki. Marker-Assisted Introgression of Quantitative Resistance Gene pi21 Confers Broad Spectrum Resistance to Rice Blast [J]. Rice Science, 2020, 27(2): 113-123. |
| [9] | Challagulla Vineela, Bhattarai Surya, J. Midmore David. In-vitro vs in-vivo Inoculation: Screening for Resistance of Australian Rice Genotypes Against Blast Fungus [J]. Rice Science, 2015, 22(3): 132-137. |
| [10] | Y J P K MITHRASENA1, W S S WIJESUNDERA2, R L C WIJESUNDERA3, D C WIMALASIRI2, R P N PRIYANTHI2. Pathogenic and Genetic Diversity of Magneporthe oryzae Populations from Sri Lanka [J]. RICE SCIENCE, 2012, 19(3): 241-246. |
| [11] | CHEN De-xi, CHEN Xue-wei, LEI Cai-lin, MA Bing-tian, WANG Yu-ping, LI Shi-gui. Rice Blast Resistance of Transgenic Rice Plants with Pi-d2 Gene [J]. RICE SCIENCE, 2010, 17(3): 179-184 . |
| [12] | CHEN De-xi, CHEN Xue-wei, MA Bing-tian, WANG Yu-ping, ZHU Li-huang, LI Shi-gui, . Genetic Transformation of Rice with Pi-d2 Gene Enhances Resistance to Rice Blast Fungus Magnaporthe oryzae [J]. RICE SCIENCE, 2010, 17(1): 19-27 . |
| [13] | YANG Jian-yuan, CHEN Shen, ZENG Lie-xian, LI Yi-long, CHEN Zhen, LI Chuan-ying, ZHU Xiao-yuan. Race Specificity of Major Rice Blast Resistance Genes to Magnaporthe grisea Isolates Collected from indica Rice in Guangdong, China [J]. RICE SCIENCE, 2008, 15(4): 311-318 . |
| [14] | YI Zi-li, WANG Zi-xuan, QIN Jing-ping, JIANG Jian-xiong, TAN Yan-ning, ZHOU Qing-ming. Transfer of Lysozyme Gene into indica Parents of Hybrid Rice by Backcrossing [J]. RICE SCIENCE, 2006, 13(4): 234-242 . |
| [15] | LIU Er-ming , XIAO Yi-long , YI You-jin , ZHUANG Jie-yun , ZHENG Kang-le , LOU Feng . Relationship Between Blast Resistance Phenotypes and Resistance Gene Analogue Profiles in Rice [J]. RICE SCIENCE, 2005, 12(2): 75-82 . |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||