
Rice Science ›› 2023, Vol. 30 ›› Issue (6): 567-576.DOI: 10.1016/j.rsci.2023.07.001
• Research Papers • Previous Articles Next Articles
Xia Xiaodong1,2,#, Zhang Xiaobo2,#, Wang Zhonghao2, Cheng Benyi2, Sun Huifeng3, Xu Xia2, Gong Junyi2, Yang Shihua2, Wu Jianli2, Shi Yongfeng2( ), Xu Rugen1(
), Xu Rugen1( )
)
Received:2023-04-12
Accepted:2023-07-15
Online:2023-11-28
Published:2023-08-10
Contact:
Xu Rugen (rgxu@yzu.edu.cn);
Shi Yongfeng (shiyongfeng@caas.cn)
About author:#These authors contributed equally to this work
Xia Xiaodong, Zhang Xiaobo, Wang Zhonghao, Cheng Benyi, Sun Huifeng, Xu Xia, Gong Junyi, Yang Shihua, Wu Jianli, Shi Yongfeng, Xu Rugen. Mapping and Functional Analysis of LE Gene in a Lethal Etiolated Rice Mutant at Seedling Stage[J]. Rice Science, 2023, 30(6): 567-576.
Add to citation manager EndNote|Ris|BibTeX
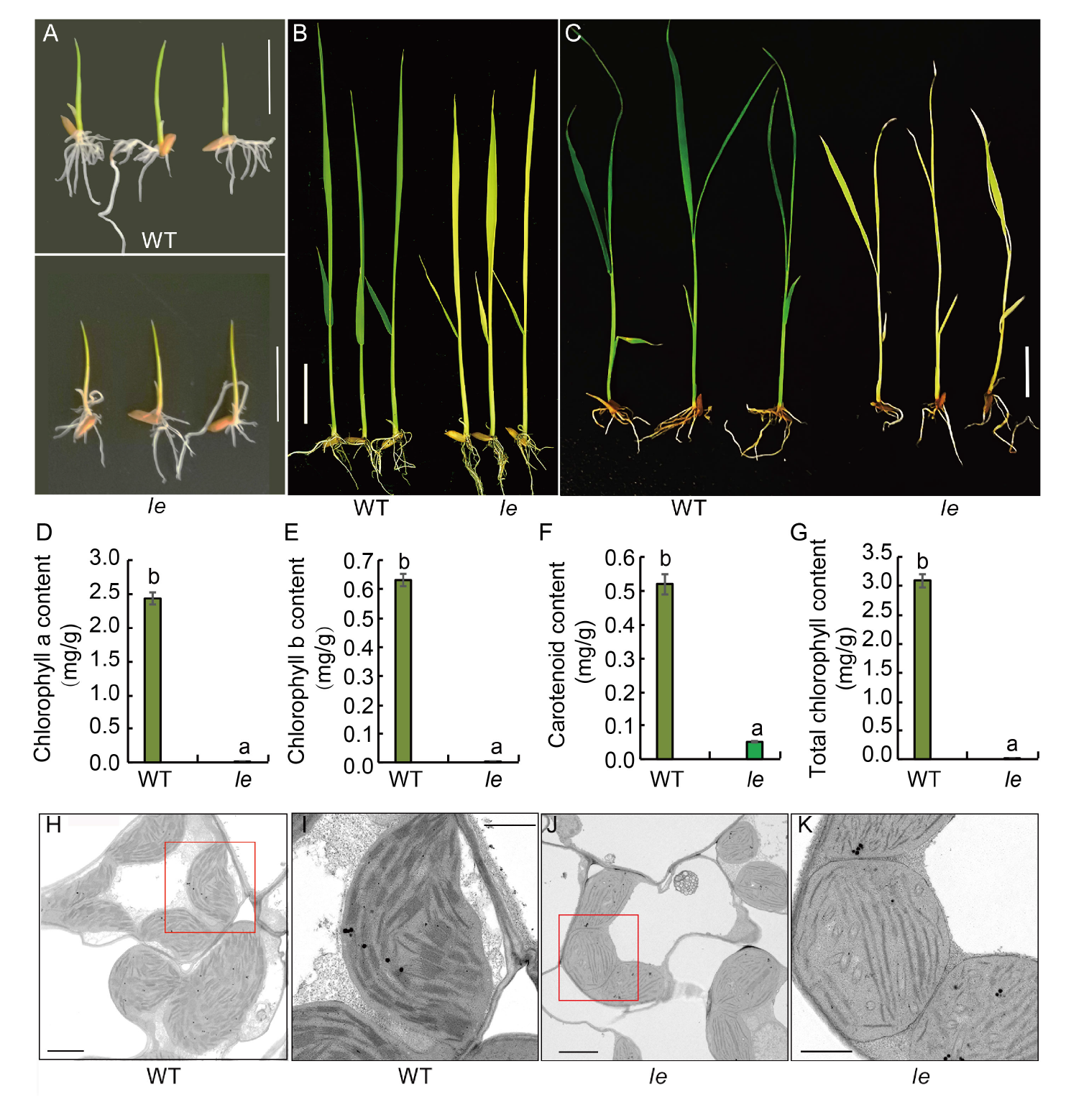
Fig. 1. Phenotype, chlorophyll content and ultrastructure of chloroplasts in wild type (WT) and le mutant. A?C, Phenotypes of WT and le mutant plants at 3 (A), 7 (B) and 14 (C) d after sowing. Scale bars, 2 cm. D?G, Contents of chlorophyll a (D), chlorophyll b (E), carotenoids (F) and total chlorophyll (G) in the mutant le and its WT. Data are Mean ± SD (n = 3). Different lowercase letters above the bars indicate statistical significances at the 0.05 probability level. H?K, Chloroplast ultrastructure of WT (H) and mutant le (J), and their enlarged parts highlighted by red boxes (I and K). Scale bars are 2 μm in H and J, and 1 μm in I and K.
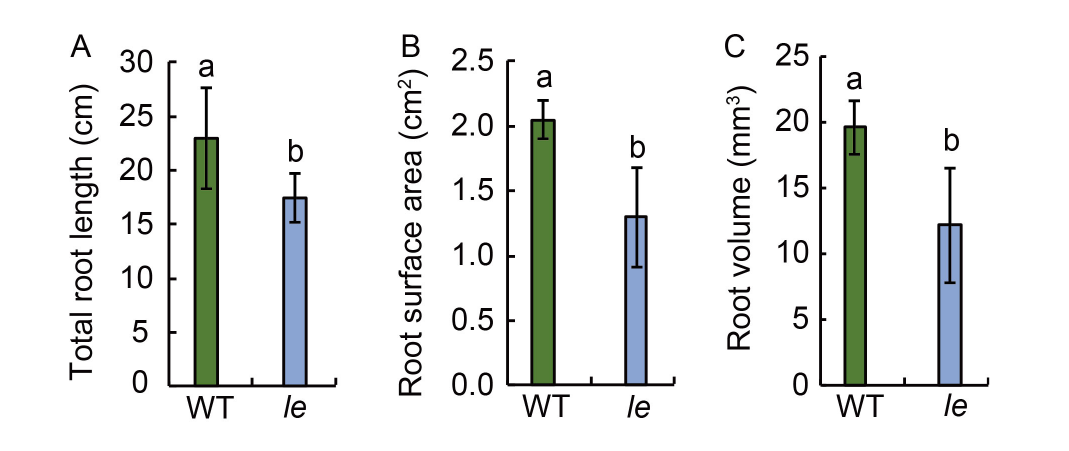
Fig. 2. Total root length (A), root surface area (B), and root volume (C) of seedlings of mutant le and its wild type (WT). Data are Mean ± SD (n = 3). Different lowercase letters above the bars indicate statistical significances at the 0.05 probability level.
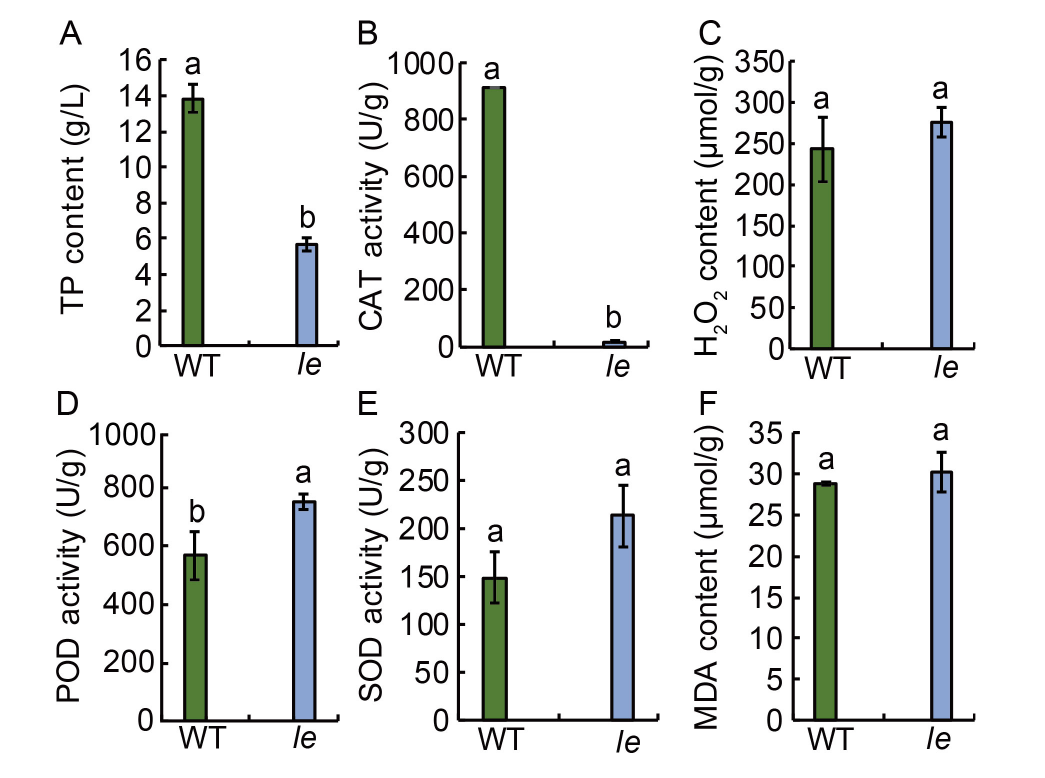
Fig. 3. Physiological and biochemical parameters of mutant le and its wild type (WT). A?F, Total intracellular protein (TP) content (A), catalase (CAT) activity (B), hydrogen peroxide (H2O2) content (C), peroxidase (POD) activity (D), superoxide dismutase (SOD) activity (E), and malondialdehyde (MDA) content (F) in le and WT. Data are Mean ± SD (n = 3). Different lowercase letters above the bars indicate statistical significances at the 0.05 probability level.
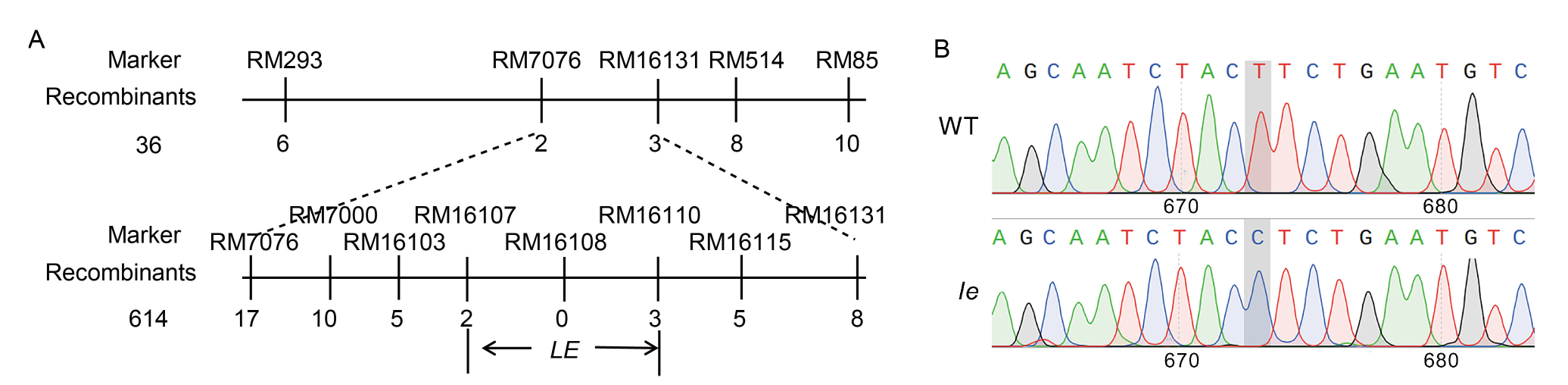
Fig. 4. Fine mapping of LE gene conferring lethal etiolated mutation (A) and differences in gene sequence of mutant le and its wild type (WT) at LOC_Os03g59640 (B).
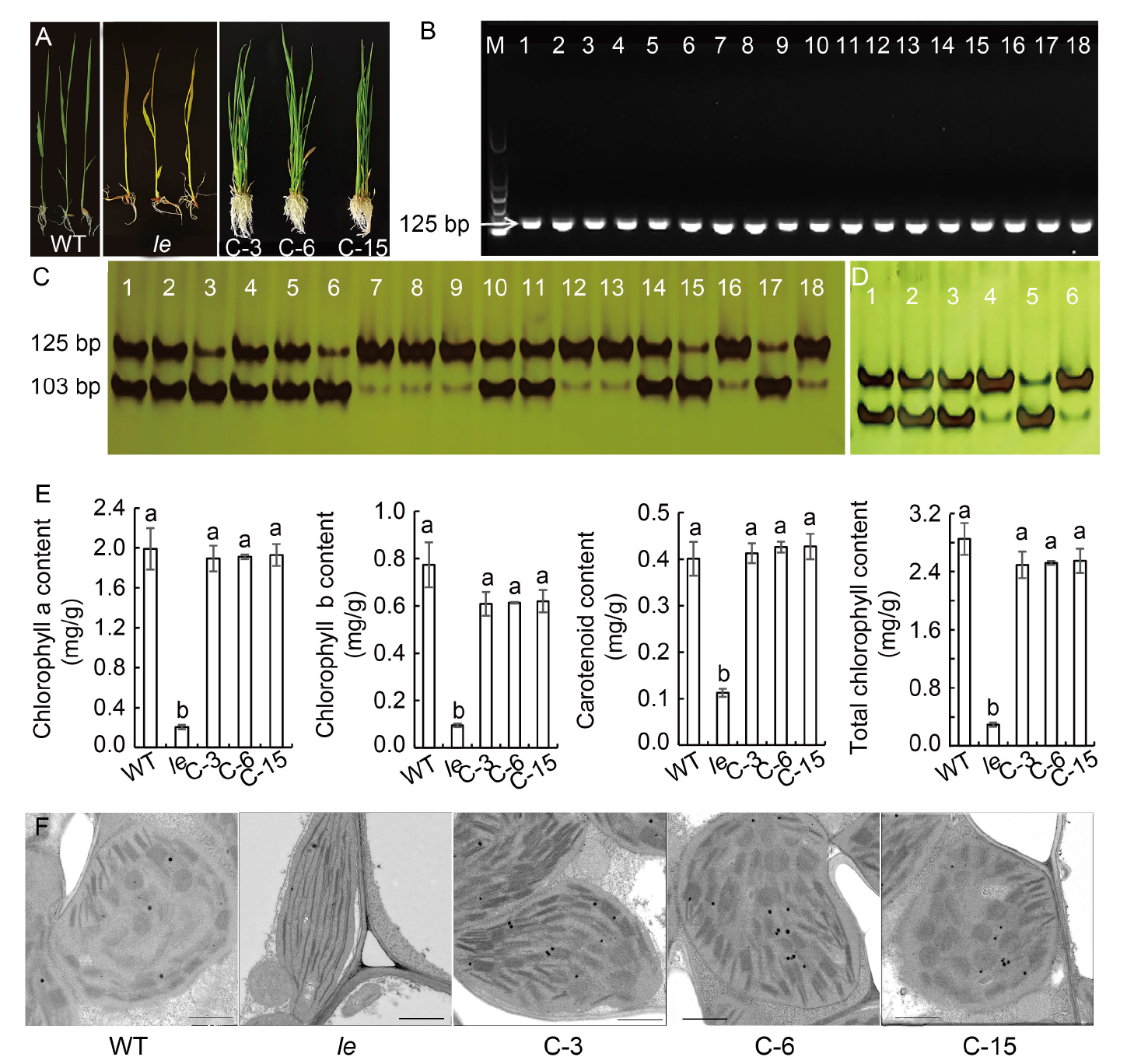
Fig. 5. Verification, phenotypes, chlo- rophyll contents, and ultrastructure of chloroplasts of complementary plants. A, Phenotypes of wild type (WT), le mutant and complementary plants (C-3, C-6, and C-15). B, PCR amplification results with primer D-1 using a 739-bp amplification product as template. M, Marker; Lanes 1?15, Transformed plants (C1?C15); Lane 16, WT; Lane 17, le mutant; Lane 18, HM133 mutant. C, Digestion results of primer D-1-based amplification products by Kpn I. Lanes 1?15, Transformed plants (C1?C15); Lane 16, WT; Lane 17, le mutant; Lane 18, HM133 mutant. D, Digestion results of transformants by Kpn I. Lanes 1?6 represent C-3, C-6, C-15, WT, le, and HM133, respectively. E, Contents of chlorophyll a, chlorophyll b, carotenoids, and total chlorophyll. Data are Mean ± SD (n = 3). Different lowercase letters above the bars indicate statistical significances at the 0.05 probability level. F, Chloroplast ultrastructures of WT, le, C-3, C-6, and C-15. Scale bars, 1 μm.
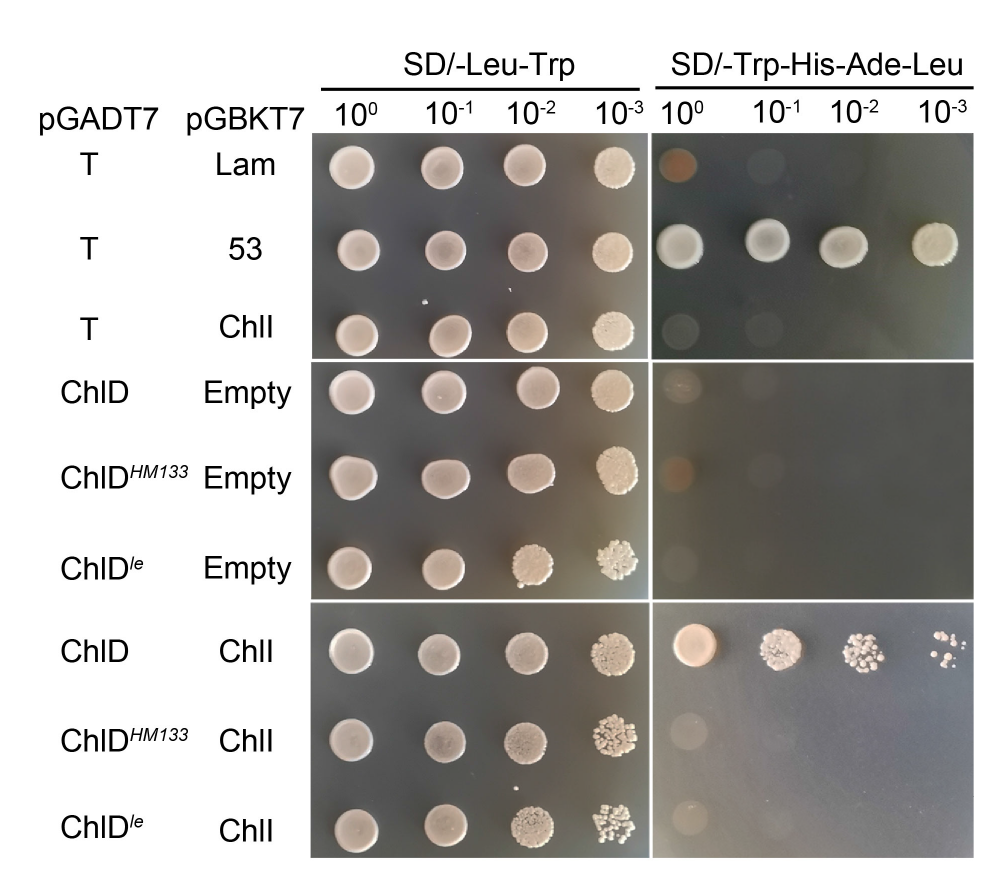
Fig. 6. Interaction of ChlDle and ChlI detected by yeast two-hybrid assay. The interaction of pGBKT7Lam and pGADT7T is used as negative control, and the interaction of pGBKT753 and pGADT7T is used as positive controls.
| [1] | Adams N B P, Vasilev C, Brindley A A, Hunter C N. 2016. Nanomechanical and thermophoretic analyses of the nucleotide- dependent interactions between the AAA+ subunits of Mg chelatase. J Am Chem Soc, 138(20): 6591-6597. |
| [2] | Brindley A A, Adams N B P, Hunter C N, Reid J D. 2015. Five glutamic acid residues in the C-terminal domain of the ChlD subunit play a major role in conferring Mg2+ cooperativity upon magnesium chelatase. Biochemistry, 54(44): 6659-6662. |
| [3] | Chen J H, Chen S T, He N Y, Wang Q L, Zhao Y, Gao W, Guo F Q. 2020. Nuclear-encoded synthesis of the D1 subunit of photosystem II increases photosynthetic efficiency and crop yield. Nat Plants, 6(5): 570-580. |
| [4] | Deng X J, Zhang H Q, Wang Y, He F, Liu J L, Xiao X, Shu Z F, Li W, Wang G H, Wang G L. 2014. Mapped clone and functional analysis of leaf-color gene Ygl7 in a rice hybrid (Oryza sativa L. ssp. indica). PLoS One, 9(6): e99564. |
| [5] | Farmer D A, Brindley A A, Hitchcock A, Jackson P J, Johnson B, Dickman M J, Hunter C N, Reid J D, Adams N B P. 2019. The ChlD subunit links the motor and porphyrin binding subunits of magnesium chelatase. Biochem J, 476(13): 1875-1887. |
| [6] | Gibson L C D, Jensen P E, Hunter C N. 1999. Magnesium chelatase from Rhodobacter sphaeroides: Initial characterization of the enzyme using purified subunits and evidence for a BchI-BchD complex. Biochem J, 337(2): 243. |
| [7] | He J Y, Wang Y Y, Ren Y F, Zhou G Q, Yang L J. 2009. Effect of cadmium on root morphology and physiological characteristics of rice seedlings. Ecol Environ Sci, 18(5): 1863-1868. (in Chinese with English abstract) |
| [8] | Ishijima S, Uchibori A, Takagi H, Maki R, Ohnishi M. 2003. Light- induced increase in free Mg2+ concentration in spinach chloroplasts: Measurement of free Mg2+ by using a fluorescent probe and necessity of stromal alkalinization. Arch Biochem Biophys, 412(1): 126-132. |
| [9] | Jensen P E, Gibson L C D, Henningsen K W, Hunter C N. 1996. Expression of the chlI, chlD, and chlH genes from the Cyanobacterium synechocystis PCC6803 in Escherichia coli and demonstration that the three cognate proteins are required for magnesium-protoporphyrin chelatase activity. J Biol Chem, 271(28): 16662-16667. |
| [10] | Jensen P E, Gibson L C, Hunter C N. 1999. ATPase activity associated with the magnesium-protoporphyrin IX chelatase enzyme of Synechocystis PCC6803: Evidence for ATP hydrolysis during Mg2+ insertion, and the MgATP-dependent interaction of the ChlI and ChlD subunits. Biochem J, 339: 127-134. |
| [11] | Kang S J, Fang Y X, Zou G X, Ruan B P, Zhao J A, Dong G J, Yan M X, Gao Z Y, Zhu L. 2015. White-green leaf gene encoding protochlorophyllide oxidoreductase B is involved in chlorophyll synthesis of rice. Crop Sci, 55(1): 284-293. |
| [12] | Lake V, Olsson U, Willows R D, Hansson M. 2004. ATPase activity of magnesium chelatase subunit I is required to maintain subunit D in vivo. Eur J Biochem, 271(11): 2182-2188. |
| [13] | Lee S, Kim J H, Yoo E S, Lee C H, Hirochika H, An G. 2005. Differential regulation of chlorophyll a oxygenase genes in rice. Plant Mol Biol, 57(6): 805-818. |
| [14] | Lundqvist J, Elmlund H, Wulff R P, Berglund L, Elmlund D, Emanuelsson C, Hebert H, Willows R D, Hansson M, Lindahl M, Al-Karadaghi S. 2010. ATP-induced conformational dynamics in the AAA+ motor unit of magnesium chelatase. Structure, 18(3): 354-365. |
| [15] | Luo S, Luo T, Peng P, Li Y P, Li X G. 2015. Activity regulation and functions of Mg chelatase. Plant Physiol J, 51(6): 806-812. (in Chinese with English abstract) |
| [16] | Luo S, Luo T, Liu Y N, Li Z W, Fan S Y, Wu C J. 2018. N-terminus plus linker domain of Mg-chelatase D subunit is essential for Mg-chelatase activity in Oryza sativa. Biochem Biophys Res Commun, 497(2): 749-755. |
| [17] | Luo T, Fan T T, Liu Y N, Rothbart M, Yu J, Zhou S X, Grimm B, Luo M Z. 2012. Thioredoxin redox regulates ATPase activity of magnesium chelatase CHLI subunit and modulates redox- mediated signaling in tetrapyrrole biosynthesis and homeostasis of reactive oxygen species in pea plants. Plant Physiol, 159(1): 118-130. |
| [18] | Luo T, Luo S, Araújo W L, Schlicke H, Rothbart M, Yu J, Fan T T, Fernie A R, Grimm B, Luo M Z. 2013. Virus-induced gene silencing of pea CHLI and CHLD affects tetrapyrrole biosynthesis, chloroplast development and the primary metabolic network. Plant Physiol Biochem, 65: 17-26. |
| [19] | Lv X G, Shi Y F, Xu X A, Wei Y L, Wang H M, Zhang X B, Wu J L. 2015. Oryza sativa chloroplast signal recognition particle 43 (OscpSRP43) is required for chloroplast development and photosynthesis. PLoS One, 10(11): e0143249. |
| [20] | Masuda T. 2008. Recent overview of the Mg branch of the tetrapyrrole biosynthesis leading to chlorophylls. Photosynth Res, 96(2): 121-143. |
| [21] | Papenbrock J, Mock H P, Kruse E, Grimm B. 1999. Expression studies in tetrapyrrole biosynthesis: Inverse maxima of magnesium chelatase and ferrochelatase activity during cyclic photoperiods. Planta, 208(2): 264-273. |
| [22] | Reid J D, Hunter C N. 2002. Current understanding of the function of magnesium chelatase. Biochem Soc Trans, 30(4): 643-645. |
| [23] | Ruan B P, Gao Z Y, Zhao J, Zhang B, Zhang A P, Hong K, Yang S L, Jiang H Z, Liu C L, Chen G, Peng Y L, Dong G J, Guo L B, Xu Z J, Qian Q. 2017. The rice YGL gene encoding an Mg2+- chelatase ChlD subunit is affected by temperature for chlorophyll biosynthesis. J Plant Biol, 60(4): 314-321. |
| [24] | Sakuraba Y, Rahman M L, Cho S H, Kim Y S, Koh H J, Yoo S C, Paek N C. 2013. The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions. Plant J, 74(1): 122-133. |
| [25] | Sawicki A, Zhou S X, Kwiatkowski K, Luo M Z, Willows R D. 2017. 1-N-histidine phosphorylation of ChlD by the AAA+ ChlI2 stimulates magnesium chelatase activity in chlorophyll synthesis. Biochem J, 474(12): 2095-2105. |
| [26] | Shang H H. 2022. Gene isolation and functional analysis of a spotted-leaf mutant spl26in rice. Wuhan, China: Huazhong Agricultural University. (in Chinese with English abstract) |
| [27] | Shi Y F, He Y, Guo D, Lv X G, Huang Q N, Wu J L. 2016. Genetic analysis and gene mapping of a pale green leaf mutant HM133 in rice. Chin J Rice Sci, 30(6): 603-610. (in Chinese with English abstract) |
| [28] | Sun X Q, Wang B, Xiao Y H, Wan C M, Deng X J, Wang P R. 2011. Genetic analysis and fine mapping of gene ygl98 for yellow-green leaf of rice. Acta Agron Sin, 37(6): 991-997. (in Chinese with English abstract) |
| [29] | Walker C J, Willows R D. 1997. Mechanism and regulation of Mg-chelatase. Biochem J, 327(Pt 2): 321-333. |
| [30] | Wang P R, Gao J X, Wan C M, Zhang F T, Xu Z J, Huang X Q, Sun X Q, Deng X J. 2010. Divinyl chlorophyll(ide) a can be converted to monovinyl chlorophyll(ide) a by a divinyl reductase in rice. Plant Physiol, 153(3): 994-1003. |
| [31] | Willows R D, Gibson L C D, Kanangara C G, Hunter C N, von Wettstein D. 1996. Three separate proteins constitute the magnesium chelatase of Rhodobacter sphaeroides. Eur J Biochem, 235(1/2): 438-443. |
| [32] | Wu Z M, Zhang X, He B, Diao L P, Sheng S L, Wang J L, Guo X P, Su N, Wang L F, Jiang L, Wang C M, Zhai H Q, Wan J M. 2007. A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol, 145(1): 29-40. |
| [33] | Zhang H, Liu L L, Cai M H, Zhu S S, Zhao J Y, Zheng T H, Xu X Y, Zeng Z Q, Niu J, Jiang L, Chen S H, Wan J M. 2015. A point mutation of magnesium chelatase OsCHLI gene dampens the interaction between CHLI and CHLD subunits in rice. Plant Mol Biol Rep, 33(6): 1975-1987. |
| [34] | Zhang H T, Li J J, Yoo J H, Yoo S C, Cho S H, Koh H J, Seo H S, Paek N C. 2006. Rice Chlorina-1 and Chlorina-9 encode ChlD and ChlI subunits of Mg-chelatase, a key enzyme for chlorophyll synthesis and chloroplast development. Plant Mol Biol, 62(3): 325-337. |
| [35] | Zhang T, Feng P, Li Y F, Yu P, Yu G L, Sang X C, Ling Y H, Zeng X Q, Li Y D, Huang J Y, Zhang T Q, Zhao F M, Wang N, Zhang C W, Yang Z L, Wu R H, He G H. 2018. VIRESCENT- ALBINO LEAF 1 regulates leaf colour development and cell division in rice. J Exp Bot, 69(20): 4791-4804. |
| [36] | Zhou S X, Sawicki A, Willows R D, Luo M Z. 2012. C-terminal residues of Oryza sativa GUN4 are required for the activation of the ChlH subunit of magnesium chelatase in chlorophyll synthesis. FEBS Lett, 586(3): 205-210. |
| [1] | Tan Jingyi, Zhang Xiaobo, Shang Huihui, Li Panpan, Wang Zhonghao, Liao Xinwei, Xu Xia, Yang Shihua, Gong Junyi, Wu Jianli. ORYZA SATIVA SPOTTED-LEAF 41 (OsSPL41) Negatively Regulates Plant Immunity in Rice [J]. Rice Science, 2023, 30(5): 426-436. |
| [2] | Md. Dhin Islam, Adam H. Price, Paul D. Hallett. Effects of Root Growth of Deep and Shallow Rooting Rice Cultivars in Compacted Paddy Soils on Subsequent Rice Growth [J]. Rice Science, 2023, 30(5): 459-472. |
| [3] | Jiang Changjie, Liang Zhengwei, Xie Xianzhi. Priming for Saline-Alkaline Tolerance in Rice: Current Knowledge and Future Challenges [J]. Rice Science, 2023, 30(5): 417-425. |
| [4] | Sheikh Faruk Ahmed, Hayat Ullah, May Zun Aung, Rujira Tisarum, Suriyan Cha-Um, Avishek Datta. Iron Toxicity Tolerance of Rice Genotypes in Relation to Growth, Yield and Physiochemical Characters [J]. Rice Science, 2023, 30(4): 321-334. |
| [5] | Yousef Alhaj Hamoud, Hiba Shaghaleh, Wang Ruke, Willy Franz Gouertoumbo, Amar Ali Adam hamad, Mohamed Salah Sheteiwy, Wang Zhenchang, Guo Xiangping. Wheat Straw Burial Improves Physiological Traits, Yield and Grain Quality of Rice by Regulating Antioxidant System and Nitrogen Assimilation Enzymes under Alternate Wetting and Drying Irrigation [J]. Rice Science, 2022, 29(5): 473-488. |
| [6] | Wang Chenjiaozi, Zhao Mei, Shu Canwei, Zhou Erxun. Three Genes Related to Trehalose Metabolism Affect Sclerotial Development of Rhizoctonia solani AG-1 IA, Causal Agent of Rice Sheath Blight [J]. Rice Science, 2022, 29(3): 268-276. |
| [7] | Hyeran Moon, Young-Ah Kim, Ryoung Shin, Chang-Jin Park. Nucleus-Encoded Thylakoid Protein, OsY3IP1, Confers Enhanced Tolerance to Saline and Alkaline Stresses in Rice [J]. Rice Science, 2022, 29(3): 225-236. |
| [8] | Chen Wei, Cai Yicong, Shakeel Ahmad, Wang Yakun, An Ruihu, Tang Shengjia, Guo Naihui, Wei Xiangjin, Tang Shaoqing, Shao Gaoneng, Jiao Guiai, Xie Lihong, Hu Shikai, Sheng Zhonghua, Hu Peisong. NRL3 Interacts with OsK4 to Regulate Heading Date in Rice [J]. Rice Science, 2022, 29(3): 237-246. |
| [9] | Singh Priyanka, Pokharia Chitra, Shah Kavita. Exogenous Peroxidase Mitigates Cadmium Toxicity, Enhances Rhizobial Population and Lowers Root Knot Formation in Rice Seedlings [J]. Rice Science, 2021, 28(2): 166-177. |
| [10] | Shuting Yuan, Chunjue Xu, Wei Yan, Zhenyi Chang, Xingwang Deng, Zhufeng Chen, Jianxin Wu, Xiaoyan Tang. Alternative Splicing of OsRAD1 Defines C-Terminal Domain Essential for Protein Function in Meiosis [J]. Rice Science, 2020, 27(4): 289-301. |
| [11] | Vijayaraghavareddy Preethi, Xinyou Yin, C. Struik Paul, Makarla Udayakumar, Sreeman Sheshshayee. Responses of Lowland, Upland and Aerobic Rice Genotypes to Water Limitation During Different Phases [J]. Rice Science, 2020, 27(4): 345-354. |
| [12] | Hussain Kashif, Yingxing Zhang, Anley Workie, Riaz Aamir, Abbas Adil, Hasanuzzaman Rani Md., Hong Wang, Xihong Shen, Liyong Cao, Shihua Cheng. Association Mapping of Quantitative Trait Loci for Grain Size in Introgression Line Derived from Oryza rufipogon [J]. Rice Science, 2020, 27(3): 246-254. |
| [13] | Ting Chen, Zheng Chen, Prakash Sathe Atul, Zhihong Zhang, Liangjian Li, Huihui Shang, Shaoqing Tang, Xiaobo Zhang, Jianli Wu. Characterization of a Novel Gain-of-Function Spotted-Leaf Mutant with Enhanced Disease Resistance in Rice [J]. Rice Science, 2019, 26(6): 372-383. |
| [14] | Nurdiani Dini, Widyajayantie Dwi, Nugroho Satya. OsSCE1 Encoding SUMO E2-Conjugating Enzyme Involves in Drought Stress Response of Oryza sativa [J]. Rice Science, 2018, 25(2): 73-81. |
| [15] | Radhesh Krishnan Subramanian, Muthuramalingam Pandiyan, Pandian Subramani, Banupriya Ramachandradoss, Chithra Gunasekar, Ramesh Manikandan. Sprouted Sorghum Extract Elicits Coleoptile Emergence, Enhances Shoot and Root Acclimatization, and Maintains Genetic Fidelity in indica Rice [J]. Rice Science, 2018, 25(2): 61-72. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||