
Rice Science ›› 2023, Vol. 30 ›› Issue (6): 577-586.DOI: 10.1016/j.rsci.2023.07.004
• Research Papers • Previous Articles Next Articles
Fan Fengfeng1,2, Cai Meng1, Luo Xiong1, Liu Manman1, Yuan Huanran1, Cheng Mingxing1, Ayaz Ahmad1, Li Nengwu1, Li Shaoqing1,2( )
)
Received:2023-04-25
Accepted:2023-07-03
Online:2023-11-28
Published:2023-08-10
Contact:
Li Shaoqing (shaoqingli@whu.edu.cn)
Fan Fengfeng, Cai Meng, Luo Xiong, Liu Manman, Yuan Huanran, Cheng Mingxing, Ayaz Ahmad, Li Nengwu, Li Shaoqing. Novel QTLs from Wild Rice Oryza longistaminata Confer Strong Tolerance to High Temperature at Seedling Stage[J]. Rice Science, 2023, 30(6): 577-586.
Add to citation manager EndNote|Ris|BibTeX
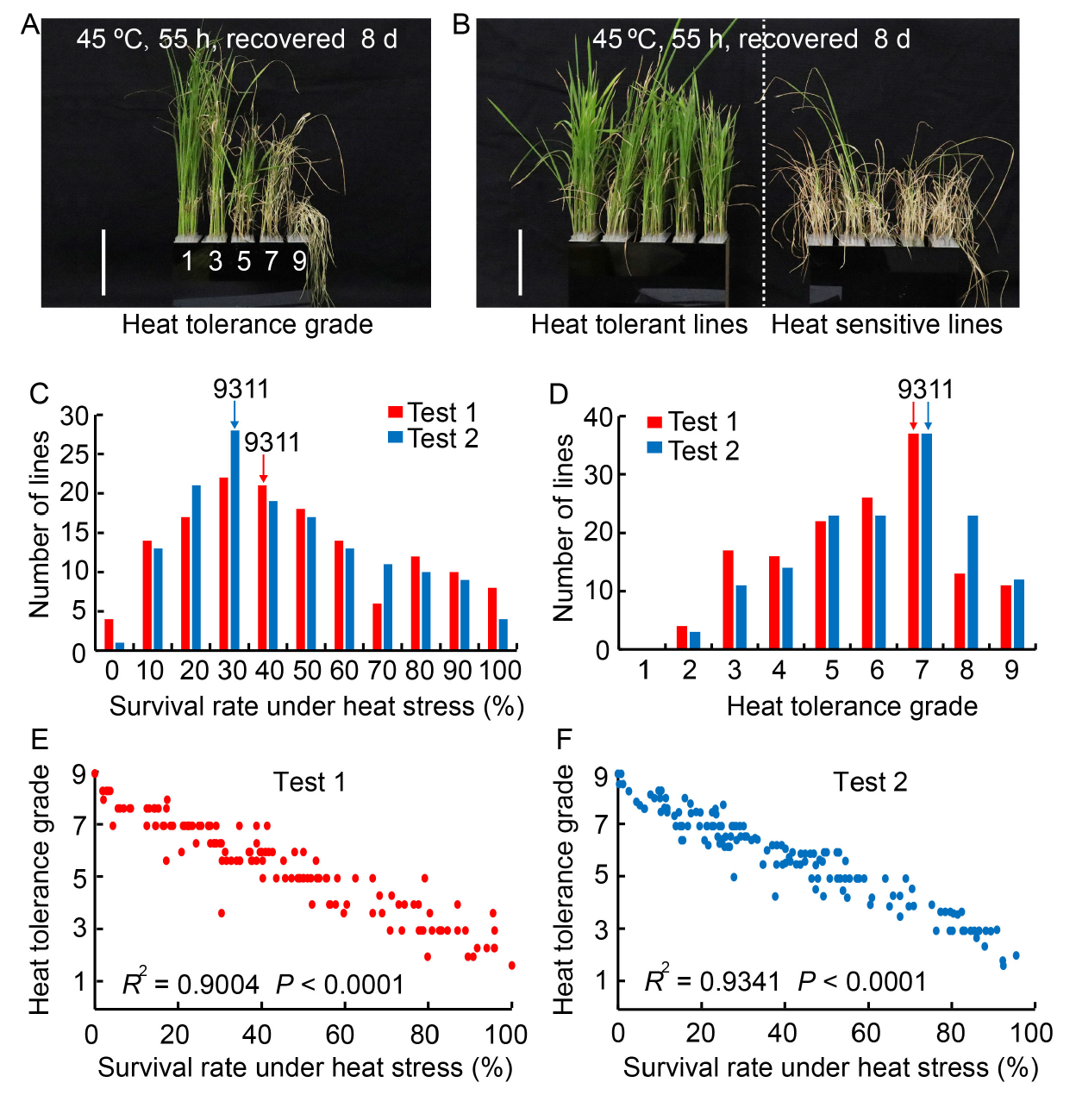
Fig. 1. Identification of heat tolerance in backcross inbred line (BIL) population and their correlations at seedling stage. A, Phenotypes correspond to heat resistance grade after high temperature treatment. Scale bar, 10 cm. B, Phenotypes of heat tolerant and heat sensitive lines after high temperature treatment. Scale bar, 10 cm. C and D, Distributions of seedling survival rate under heat stress (C) and heat tolerance grade (D) of BIL population in two tests. Three replicates were performed for each BIL in each test. E and F, Correlation between heat tolerance grade and seedling survival rate under heat stress in two tests.
| Trait | Test | 9311 | BILs | |||
|---|---|---|---|---|---|---|
| Mean ± SD | CV (%) | Range | Heritability (%) | |||
| Survival rate under heat stress (%) | Test 1 | 35.7 ± 7.5 | 41.8 ± 27.3 | 65.3 | 0.0‒100.0 | 73.59 |
| Test 2 | 28.5 ± 3.3 | 40.3 ± 25.6 | 63.5 | 0.0‒96.3 | ||
| Heat tolerance grade | Test 1 | 6.5 ± 1.0 | 5.7 ± 1.8 | 31.6 | 1.7‒9.0 | 69.86 |
| Test 2 | 7.0 ± 0.0 | 5.9 ± 1.7 | 28.8 | 1.5‒9.0 | ||
Table 1. Heat tolerance phenotypic variations in 9311 and backcross inbred lines (BILs) in two tests.
| Trait | Test | 9311 | BILs | |||
|---|---|---|---|---|---|---|
| Mean ± SD | CV (%) | Range | Heritability (%) | |||
| Survival rate under heat stress (%) | Test 1 | 35.7 ± 7.5 | 41.8 ± 27.3 | 65.3 | 0.0‒100.0 | 73.59 |
| Test 2 | 28.5 ± 3.3 | 40.3 ± 25.6 | 63.5 | 0.0‒96.3 | ||
| Heat tolerance grade | Test 1 | 6.5 ± 1.0 | 5.7 ± 1.8 | 31.6 | 1.7‒9.0 | 69.86 |
| Test 2 | 7.0 ± 0.0 | 5.9 ± 1.7 | 28.8 | 1.5‒9.0 | ||
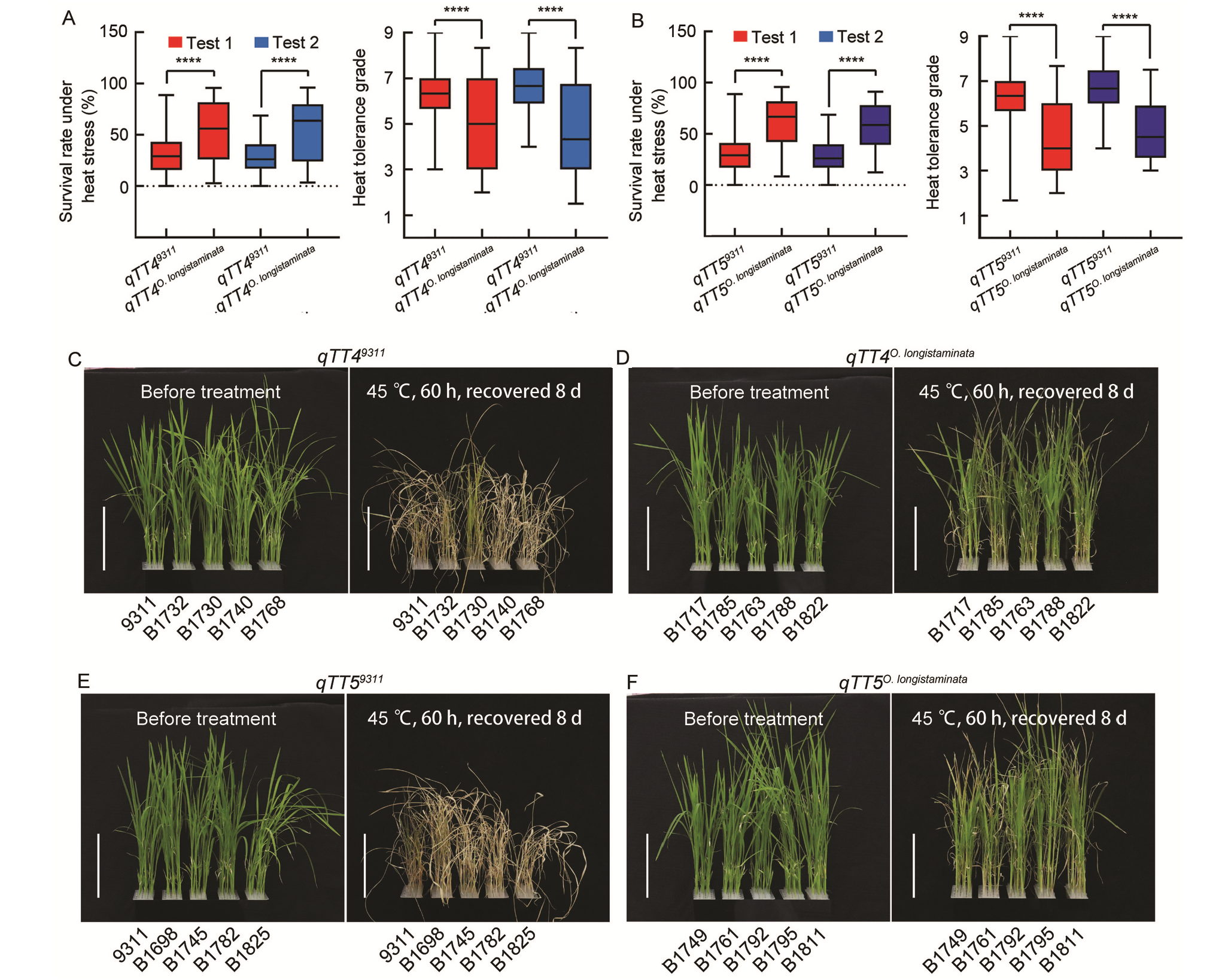
Fig. 3. Validation of function of qTT4 and qTT5. A and B, Analysis of heat tolerance of qTT4 and qTT5 in backcross inbred line population. C?F, Heat tolerance phenotypes of qTT4 (C and D) and qTT5 (E and F). Scale bars, 10 cm. qTT4(qTT5)9311 represents lines containing qTT4(qTT5) from 9311, qTT4(qTT5)O. longistaminata represents lines containing qTT4(qTT5) from O. longistaminata. **** indicates significant differences at the 0.0001 level, by the Student’s t-test.
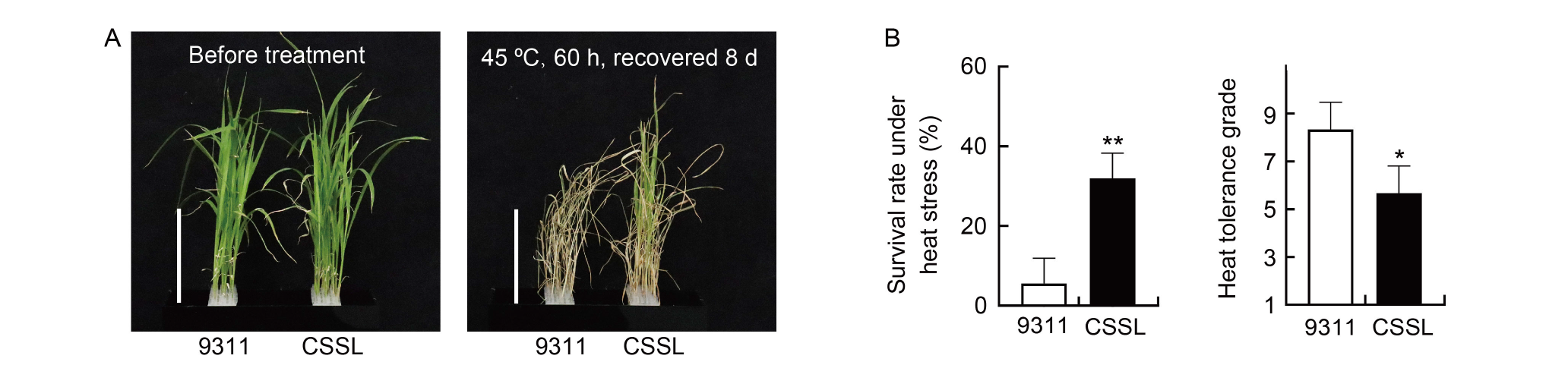
Fig. 4. qTT5 improves heat tolerance of rice at seedling stage. A, Phenotypes of heat tolerance for qTT5-carrying chromosome segment substitution line (CSSL) and its recurrent parent 9311. Scale bars, 10 cm. B, Seedling survival rate and heat tolerance grade of qTT5-carrying CSSL and 9311 under heat stress. Data are Mean ± SD (n = 3). * and ** indicate significant differences at the 0.05 and 0.01 levels, respectively, by the Student’s t-test.
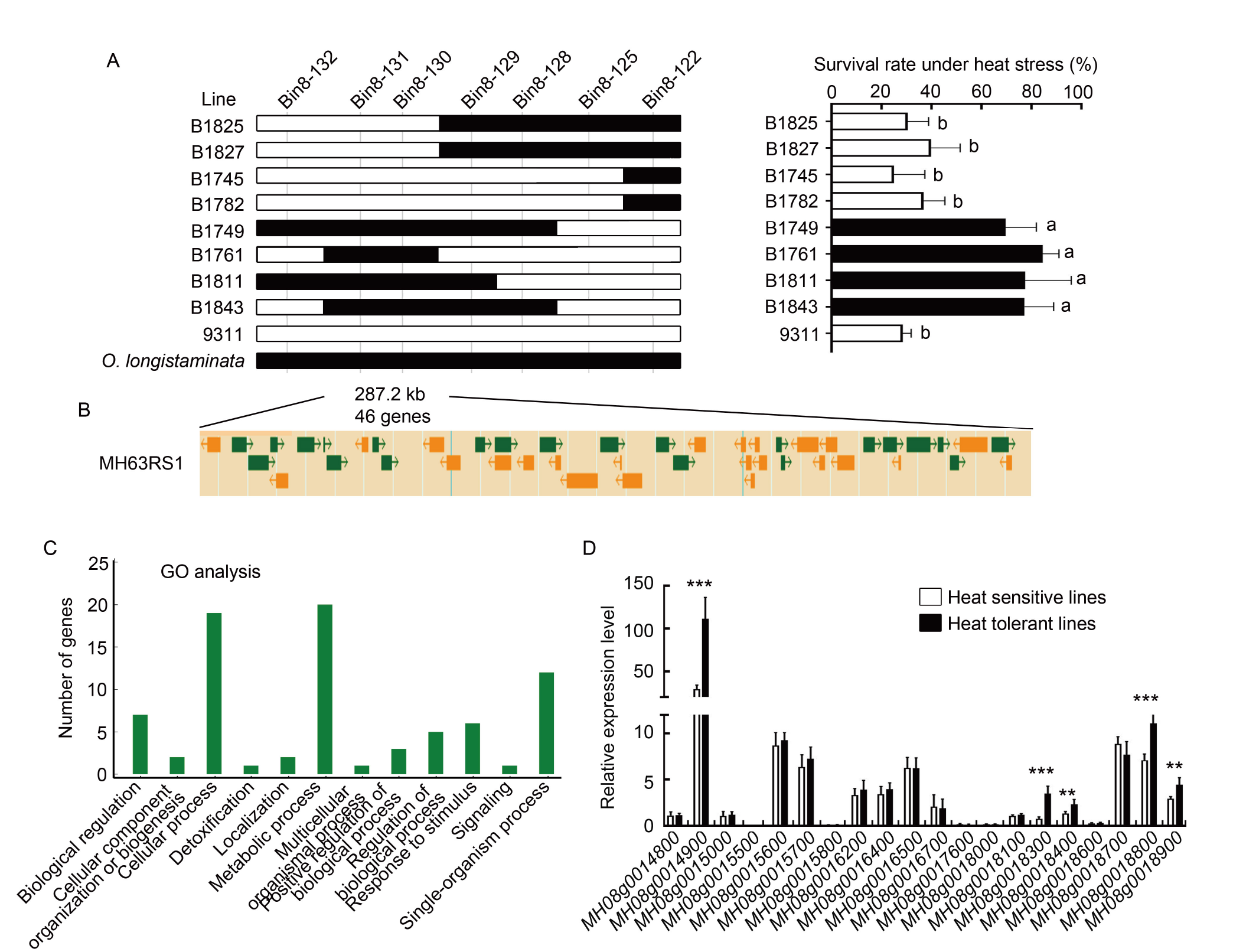
Fig. 5. Analysis of qTT5 candidate genes. A, Genotype of progeny testing for qTT5 delimited the locus to a 287.2-kb stretch flanked by Bin8-130 and Bin8-131. Seedling survival rate under heat stress was shown for recombinant backcross inbred lines and parental 9311. Data are Mean ± SD (n = 3). The different lowercase letters represent significant differences (P < 0.05) as determined by the Student’s t-test. B, A total of 46 putative genes was annotated within the localization interval. C, Gene Ontology (GO) analysis of 46 candidate genes in the localization interval. D, Expression levels of 20 genes under the GO term of metabolic process and response to stimulus. The expression levels of five heat sensitive lines (white) and five heat resistant lines (black) were calibrated to rice Ubiquitin gene expression. Date are Mean ± SD (n = 5). ** and *** indicate significant differences at the 0.01 and 0.001 levels, respectively, by the Student’s t-test.
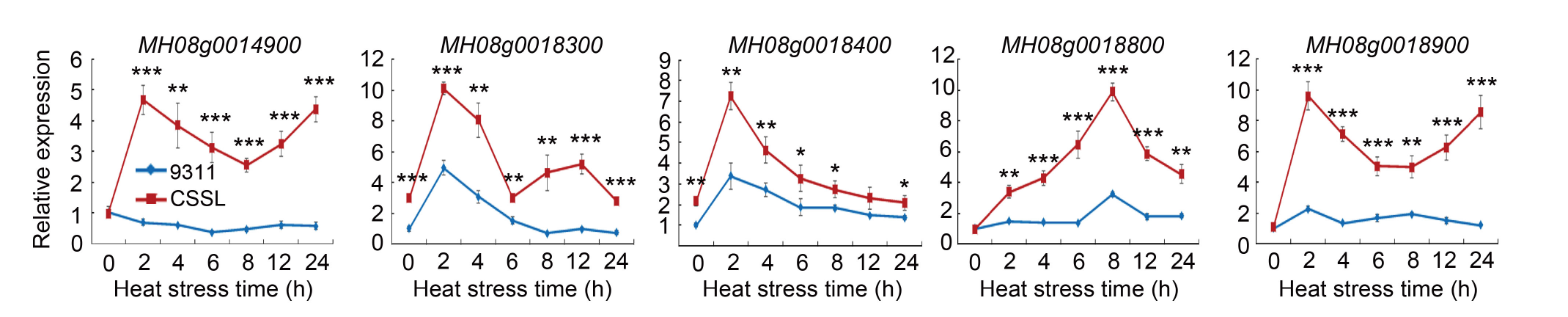
Fig. 6. Expression analysis of candidate genes in plants exposed to 45 ºC heat stress in a phytotron for different time periods. The transcript level is shown as the expression relative to the Ubiquitin gene expression. Data are Mean ± SD (n = 5). *, **, and *** indicate significant differences at the 0.05, 0.01 and 0.001 levels, respectively, by the Student’s t-test.
| [1] | Ambavaram M M R, Basu S, Krishnan A, Ramegowda V, Batlang U, Rahman L, Baisakh N, Pereira A. 2014. Coordinated regulation of photosynthesis in rice increases yield and tolerance to environmental stress. Nat Commun, 5: 5302. |
| [2] | Ballinger C A, Connell P, Wu Y X, Hu Z Y, Thompson L J, Yin L Y, Patterson C. 1999. Identification of CHIP, a novel tetratricopeptide repeat-containing protein that interacts with heat shock proteins and negatively regulates chaperone functions. Mol Cell Biol, 19(6): 4535-4545. |
| [3] | Cao Z B, Li Y, Tang H W, Zeng B H, Tang X Y, Long Q Z, Wu X F, Cai Y H, Yuan L F, Wan J L. 2020. Fine mapping of the qHTB1-1 QTL, which confers heat tolerance at the booting stage, using an Oryza rufipogon Griff. introgression line. Theor Appl Genet, 133(4): 1161-1175. |
| [4] | Dhatt B K, Paul P, Sandhu J, Hussain W, Irvin L, Zhu F Y, Adviento-Borbe M A, Lorence A, Staswick P, Yu H F, Morota G, Walia H. 2021. Allelic variation in rice Fertilization Independent Endosperm 1 contributes to grain width under high night temperature stress. New Phytol, 229(1): 335-350. |
| [5] | Fan F F, Li N W, Chen Y P, Liu X D, Sun H, Wang J, He G C, Zhu Y G, Li S Q. 2017. Development of elite BPH-resistant wide- spectrum restorer lines for three and two line hybrid rice. Front Plant Sci, 8: 986. |
| [6] | Fan F F, Long W X, Liu M M, Yuan H R, Pan G J, Li N W, Li S Q. 2019. Quantitative trait locus mapping of the combining ability for yield-related traits in wild rice Oryza longistaminata. J Agric Food Chem, 67(32): 8766-8772. |
| [7] | Hu C M, Jiang J H, Li Y L, Song S J, Zou Y, Jing C Y, Zhang Y, Wang D Z, He Q A, Dang X J. 2022. QTL mapping and identification of candidate genes using a genome-wide association study for heat tolerance at anthesis in rice (Oryza sativa L.). Front Genet, 13: 983525. |
| [8] | Huang S Y, Liu M M, Chen G L, Si F F, Fan F F, Guo Y, Yuan L, Yang F, Li S Q. 2022. Favorable QTLs from Oryza longistaminata improve rice drought resistance. BMC Plant Biol, 22(1): 136. |
| [9] | Jin J, Long W X, Wang L T, Liu X D, Pan G J, Xiang W, Li N W, Li S Q. 2018. QTL mapping of seed vigor of backcross inbred lines derived from Oryza longistaminata under artificial aging. Front Plant Sci, 9: 1909. |
| [10] | Kan Y, Lin H X. 2021. Molecular regulation and genetic control of rice thermal response. Crop J, 9(3): 497-505. |
| [11] | Kan Y, Mu X R, Zhang H, Gao J, Shan J X, Ye W W, Lin H X. 2022. TT2 controls rice thermotolerance through SCT1-dependent alteration of wax biosynthesis. Nat Plants, 8(1): 53-67. |
| [12] | Khong G N, Pati P K, Richaud F, Parizot B, Bidzinski P, Mai C D, Bès M, Bourrié I, Meynard D, Beeckman T, Selvaraj M G, Manabu I, Genga A M, Brugidou C, Nang Do V, Guiderdoni E, Morel J B, Gantet P. 2015. OsMADS26 negatively regulates resistance to pathogens and drought tolerance in rice. Plant Physiol, 169(4): 2935-2949. |
| [13] | Lander E S, Green P, Abrahamson J, Barlow A, Daly M J, Lincoln S E, Newberg L A. 1987. MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics, 1(2): 174-181. |
| [14] | Lee B H, Won S H, Lee H S, Miyao M, Chung W I, Kim I J, Jo J. 2000. Expression of the chloroplast-localized small heat shock protein by oxidative stress in rice. Gene, 245(2): 283-290. |
| [15] | Lee S, Lee D W, Lee Y, Mayer U, Stierhof Y D, Lee S, Jürgens G, Hwang I. 2009. Heat shock protein cognate 70-4 and an E3 ubiquitin ligase, CHIP, mediate plastid-destined precursor degradation through the ubiquitin-26S proteasome system in Arabidopsis. Plant Cell, 21(12): 3984-4001. |
| [16] | Li M, Yu H H, Liu K, Yang W L, Zhou B J, Gan L, Li S J, Zhang C, Yu B. 2021. Serrate-associated protein 1, a splicing-related protein, promotes miRNA biogenesis in Arabidopsis. New Phytol, 232(5): 1959-1973. |
| [17] | Li X M, Chao D Y, Wu Y, Huang X H, Chen K, Cui L G, Su L, Ye W W, Chen H, Chen H C, Dong N Q, Guo T, Shi M, Feng Q, Zhang P, Han B, Shan J X, Gao J P, Lin H X. 2015. Natural alleles of a proteasome α2 subunit gene contribute to thermotolerance and adaptation of African rice. Nat Genet, 47(7): 827-833. |
| [18] | Linares O F. 2002. African rice (Oryza glaberrima): History and future potential. Proc Natl Acad Sci USA, 99(25): 16360-16365. |
| [19] | Liu J G, Qin Q L, Zhang Z, Peng R H, Xiong A S, Chen J M, Yao Q H. 2009. OsHSF7gene in rice, Oryza sativa L., encodes a transcription factor that functions as a high temperature receptive and responsive factor. BMB Rep, 42(1): 16-21. |
| [20] | Liu J P, Zhang C C, Wei C C, Liu X, Wang M G, Yu F F, Xie Q, Tu J M. 2016. The RING finger ubiquitin E3 ligase OsHTAS enhances heat tolerance by promoting H2O2-induced stomatal closure in rice. Plant Physiol, 170(1): 429-443. |
| [21] | Liu X D, Fan F F, Liu M M, Long W X, Yu Y J, Yuan H R, Pan G J, Li N W, Li S Q, Liu J F. 2020. Quantitative trait loci mapping of mineral element contents in brown rice using backcross inbred lines derived from Oryza longistaminata. Front Plant Sci, 11: 1229. |
| [22] | Lo S F, Cheng M L, Hsing Y I C, Chen Y S, Lee K W, Hong Y F, Hsiao Y, Hsiao A S, Chen P J, Wong L I, Chen N C, Reuzeau C, Ho T H D, Yu S M. 2020. Rice Big Grain 1 promotes cell division to enhance organ development, stress tolerance and grain yield. Plant Biotechnol J, 18(9): 1969-1983. |
| [23] | Lobell D B, Schlenker W, Costa-Roberts J. 2011. Climate trends and global crop production since 1980. Science, 333: 616-620. |
| [24] | Long W X, Li N W, Jin J, Wang J, Dan D, Fan F F, Gao Z Y, Li S Q. 2023. Resequencing-based QTL mapping for yield and resistance traits reveals great potential of Oryza longistaminata in rice breeding. Crop J. DOI: 10.1016/j.cj.2023.03.017. |
| [25] | Meng L, Li H H, Zhang L Y, Wang J K. 2015. QTL IciMapping: Integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J, 3(3): 269-283. |
| [26] | Müller F, Rieu I. 2016. Acclimation to high temperature during pollen development. Plant Reprod, 29(1): 107-118. |
| [27] | Nevame A Y M, Emon R M, Malek M A, Hasan M M, Alam M A, Muharam F M, Aslani F, Rafii M Y, Ismail M R. 2018. Relationship between high temperature and formation of chalkiness and their effects on quality of rice. Biomed Res Int, 2018: 1653721. |
| [28] | Reuscher S, Furuta T, Bessho-Uehara K, Cosi M, Jena K K, Toyoda A, Fujiyama A, Kurata N, Ashikari M. 2018. Assembling the genome of the African wild rice Oryza longistaminata by exploiting synteny in closely related Oryza species. Commun Biol, 1: 162. |
| [29] | Sakai H, Ikawa H, Tanaka T, Numa H, Minami H, Fujisawa M, Shibata M, Kurita K, Kikuta A, Hamada M, Kanamori H, Namiki N, Wu J Z, Itoh T, Matsumoto T, Sasaki T. 2011. Distinct evolutionary patterns of Oryza glaberrima deciphered by genome sequencing and comparative analysis. Plant J, 66(5): 796-805. |
| [30] | Sarsu F, Ghanim A M A, Das P, Bahuguna R N, Kusolwa P M, Ashraf M, Singla-Pareek S L, Pareek A, Forster B P, Ingelbrecht I. 2018. Pre-Field Screening Protocols for Heat-Tolerant Mutants in Rice. Cham, the Netherland: Springer: 15-17. |
| [31] | Sato Y, Yokoya S. 2008. Enhanced tolerance to drought stress in transgenic rice plants overexpressing a small heat-shock protein, sHSP17.7. Plant Cell Rep, 27(2): 329-334. |
| [32] | Sharma A, Kumar V, Shahzad B, Ramakrishnan M, Sidhu G P S, Bali A S, Handa N, Kapoor D, Yadav P, Khanna K, Bakshi P, Rehman A, Kohli S K, Khan E A, Parihar R D, Yuan H W, Thukral A K, Bhardwaj R, Zheng B S. 2020. Photosynthetic response of plants under different abiotic stresses: A review. J Plant Growth Regul, 39(2): 509-531. |
| [33] | She K C, Kusano H, Koizumi K, Yamakawa H, Hakata M, Imamura T, Fukuda M, Naito N, Tsurumaki Y, Yaeshima M, Tsuge T, Matsumoto K, Kudoh M, Itoh E, Kikuchi S, Kishimoto N, Yazaki J, Ando T, Yano M, Aoyama T, Sasaki T, Satoh H, Shimada H. 2010. A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant Cell, 22(10): 3280-3294. |
| [34] | Szymańska R, Ślesak I, Orzechowska A, Kruk J. 2017. Physiological and biochemical responses to high light and temperature stress in plants. Environ Exp Bot, 139: 165-177. |
| [35] | Wang Y J, Makowski L. 2018. Fine structure of conformational ensembles in adenylate kinase. Proteins, 86(3): 332-343. |
| [36] | Xu Y F, Chu C C, Yao S G. 2021. The impact of high-temperature stress on rice: Challenges and solutions. Crop J, 9(5): 963-976. |
| [37] | Yan J Q, Wang J, Li Q T, Hwang J R, Patterson C, Zhang H. 2003. AtCHIP, a U-box-containing E3 ubiquitin ligase, plays a critical role in temperature stress tolerance in Arabidopsis. Plant Physiol, 132(2): 861-869. |
| [38] | Yang Y L, Xu J, Huang L C, Leng Y J, Dai L P, Rao Y C, Chen L, Wang Y Q, Tu Z J, Hu J, Ren D Y, Zhang G H, Zhu L, Guo L B, Qian Q, Zeng D L. 2016. PGL, encoding chlorophyllide a oxygenase 1, impacts leaf senescence and indirectly affects grain yield and quality in rice. J Exp Bot, 67(5): 1297-1310. |
| [39] | Yuan L, Zhang L C, Wei X, Wang R H, Li N N, Chen G L, Fan F F, Huang S Y, Li J X, Li S Q. 2022. Quantitative trait locus mapping of salt tolerance in wild rice Oryza longistaminata. Int J Mol Sci, 23(4): 2379. |
| [40] | Zeng D L, Tian Z X, Rao Y C, Dong G J, Yang Y L, Huang L C, Leng Y J, Xu J, Sun C, Zhang G H, Hu J, Zhu L, Gao Z Y, Hu X M, Guo L B, Xiong G S, Wang Y H, Li J Y, Qian Q. 2017. Rational design of high-yield and superior-quality rice. Nat Plants, 3: 17031. |
| [41] | Zhang H, Zhou J F, Kan Y, Shan J X, Ye W W, Dong N Q, Guo T, Xiang Y H, Yang Y B, Li Y C, Zhao H Y, Yu H X, Lu Z Q, Guo S Q, Lei J J, Liao B, Mu X R, Cao Y J, Yu J J, Lin Y S, Lin H X. 2022. A genetic module at one locus in rice protects chloroplasts to enhance thermotolerance. Science, 376: 1293-1300. |
| [42] | Zhao C, Liu B, Piao S L, Wang X H, Lobell D B, Huang Y, Huang M T, Yao Y T, Bassu S, Ciais P, Durand J L, Elliott J, Ewert F, Janssens I A, Li T, Lin E D, Liu Q, Martre P, Müller C, Peng S S, Peñuelas J, Ruane A C, Wallach D, Wang T, Wu D H, Liu Z, Zhu Y, Zhu Z C, Asseng S. 2017. Temperature increase reduces global yields of major crops in four independent estimates. Proc Natl Acad Sci USA, 114(35): 9326-9331. |
| [43] | Zhou J, Zhang Y, Qi J X, Chi Y J, Fan B F, Yu J Q, Chen Z X. 2014. E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in plant stress responses. PLoS Genet, 10(1): e1004116. |
| [44] | Zou J, Liu A L, Chen X B, Zhou X Y, Gao G F, Wang W F, Zhang X W. 2009. Expression analysis of nine rice heat shock protein genes under abiotic stresses and ABA treatment. J Plant Physiol, 166(8): 851-861. |
| [1] | Prathap V, Suresh Kumar, Nand Lal Meena, Chirag Maheshwari, Monika Dalal, Aruna Tyagi. Phosphorus Starvation Tolerance in Rice Through Combined Physiological, Biochemical, and Proteome Analyses [J]. Rice Science, 2023, 30(6): 613-631. |
| [2] | Si Fengfeng, Fan Fengfeng, Wei Xiao, He Shihao, Li Xianlong, Peng Xiaojue, Li Shaoqing. Quantitative Trait Locus Mapping of High Photosynthetic Efficiency and Biomass in Oryza longistaminata [J]. Rice Science, 2022, 29(6): 569-576. |
| [3] | Kossi Lorimpo Adjah, Maxwell Darko Asante, Aboubacar Toure, Mawuli Aziadekey, Francis Osei Amoako-Andoh, Michael Frei, Yacouba Diallo, Komi Agboka. Improvement of Rice Production under Drought Conditions in West Africa: Application of QTLs in Breeding for Drought Resistance [J]. Rice Science, 2022, 29(6): 512-521. |
| [4] | Yang Ziyi, Xu Zhijian, Yang Qingwen, Qiao Weihua. Conservation and Utilization of Genetic Resources of Wild Rice in China [J]. Rice Science, 2022, 29(3): 216-224. |
| [5] | Nie Yuanyuan, Xia Hui, Ma Xiaosong, Lou Qiaojun, Liu Yi, Zhang Anling, Cheng Liang, Yan Longan, Luo Lijun. Dissecting Genetic Basis of Deep Rooting in Dongxiang Wild Rice [J]. Rice Science, 2022, 29(3): 277-287. |
| [6] | Tan Quanya, Zhu Haitao, Liu Hui, Ni Yuerong, Wu Shengze, Luan Xin, Liu Junwei, Yang Weifeng, Yang Zifeng, Zeng Ruizhen, Liu Guifu, Wang Shaokui, Zhang Guiquan. Fine Mapping of QTLs for Stigma Exsertion Rate from Oryza glaberrima by Chromosome Segment Substitution [J]. Rice Science, 2022, 29(1): 55-66. |
| [7] | Jan Mehmood, Shah Gulmeena, Yuqing Huang, Xuejiao Liu, Peng Zheng, Hao Du, Hao Chen, Jumin Tu. Development of Heat Tolerant Two-Line Hybrid Rice Restorer Line Carrying Dominant Locus of OsHTAS [J]. Rice Science, 2021, 28(1): 99-108. |
| [8] | Weidong Qi, Hongping Chen, Zuozhen Yang, Biaolin Hu, Xiangdong Luo, Bing Ai, Yuan Luo, Yu Huang, Jiankun Xie, Fantao Zhang. Systematic Characterization of Long Non-Coding RNAs and Their Responses to Drought Stress in Dongxiang Wild Rice [J]. Rice Science, 2020, 27(1): 21-31. |
| [9] | Cheabu Sulaiman, Panichawong Nat, Rattanametta Prisana, Wasuri Boonthong, Kasemsap Poonpipope, Arikit Siwaret, Vanavichit Apichart, Malumpong Chanate. Screening for Spikelet Fertility and Validation of Heat Tolerance in a Large Rice Mutant Population [J]. Rice Science, 2019, 26(4): 229-238. |
| [10] | Cheabu Sulaiman, Moung-ngam Peerapon, Arikit Siwaret, Vanavichit Apichart, Malumpong Chanate. Effects of Heat Stress at Vegetative and Reproductive Stages on Spikelet Fertility [J]. Rice Science, 2018, 25(4): 218-226. |
| [11] | P. M. Swamy B., Kaladhar K., Anuradha K., K. Batchu Anil, Longvah T., Sarla N.. QTL Analysis for Grain Iron and Zinc Concentrations in Two O. nivara Derived Backcross Populations [J]. Rice Science, 2018, 25(4): 197-207. |
| [12] | Fantao Zhang, Yuan Luo, Meng Zhang, Yi Zhou, Hongping Chen, Biaolin Hu, Jiankun Xie. Identification and Characterization of Drought Stress- Responsive Novel microRNAs in Dongxiang Wild Rice [J]. Rice Science, 2018, 25(4): 175-184. |
| [13] | Yaobin Qin, Peng Cheng, Yichen Cheng, Yue Feng, Derun Huang, Tingxu Huang, Xianjun Song, Jiezheng Ying. QTL-Seq Identified a Major QTL for Grain Length and Weight in Rice Using Near Isogenic F2 Population [J]. Rice Science, 2018, 25(3): 121-131. |
| [14] | Haritha G., P. M. Swamy B., L. Naik M., Jyothi B., Divya B., Malathi S., Sarla N.. Yield Traits and Associated Marker Segregation in Elite Introgression Lines Derived from O. sativa × O. nivara [J]. Rice Science, 2018, 25(1): 19-31. |
| [15] | Vivitha P., Raveendran M., Vijayalakshmi D.. Introgression of QTLs Controlling Spikelet Fertility Maintains Membrane Integrity and Grain Yield in Improved White Ponni Derived Progenies Exposed to Heat Stress [J]. Rice Science, 2017, 24(1): 32-40. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||