
Rice Science ›› 2023, Vol. 30 ›› Issue (6): 587-597.DOI: 10.1016/j.rsci.2023.09.001
• Research Papers • Previous Articles Next Articles
Serena Reggi1, Elisabetta Onelli2, Alessandra Moscatelli2( ), Nadia Stroppa2, Matteo Dell’Anno1, Kiril Perfanov3, Luciana Rossi1
), Nadia Stroppa2, Matteo Dell’Anno1, Kiril Perfanov3, Luciana Rossi1
Received:2023-01-21
Accepted:2023-06-19
Online:2023-11-28
Published:2023-08-10
Contact:
Alessandra Moscatelli (alessandra.moscatelli@unimi.it)
Serena Reggi, Elisabetta Onelli, Alessandra Moscatelli, Nadia Stroppa, Matteo Dell’Anno, Kiril Perfanov, Luciana Rossi. Seed-Specific Expression of Apolipoprotein A-IMilano Dimer in Engineered Rice Lines[J]. Rice Science, 2023, 30(6): 587-597.
Add to citation manager EndNote|Ris|BibTeX
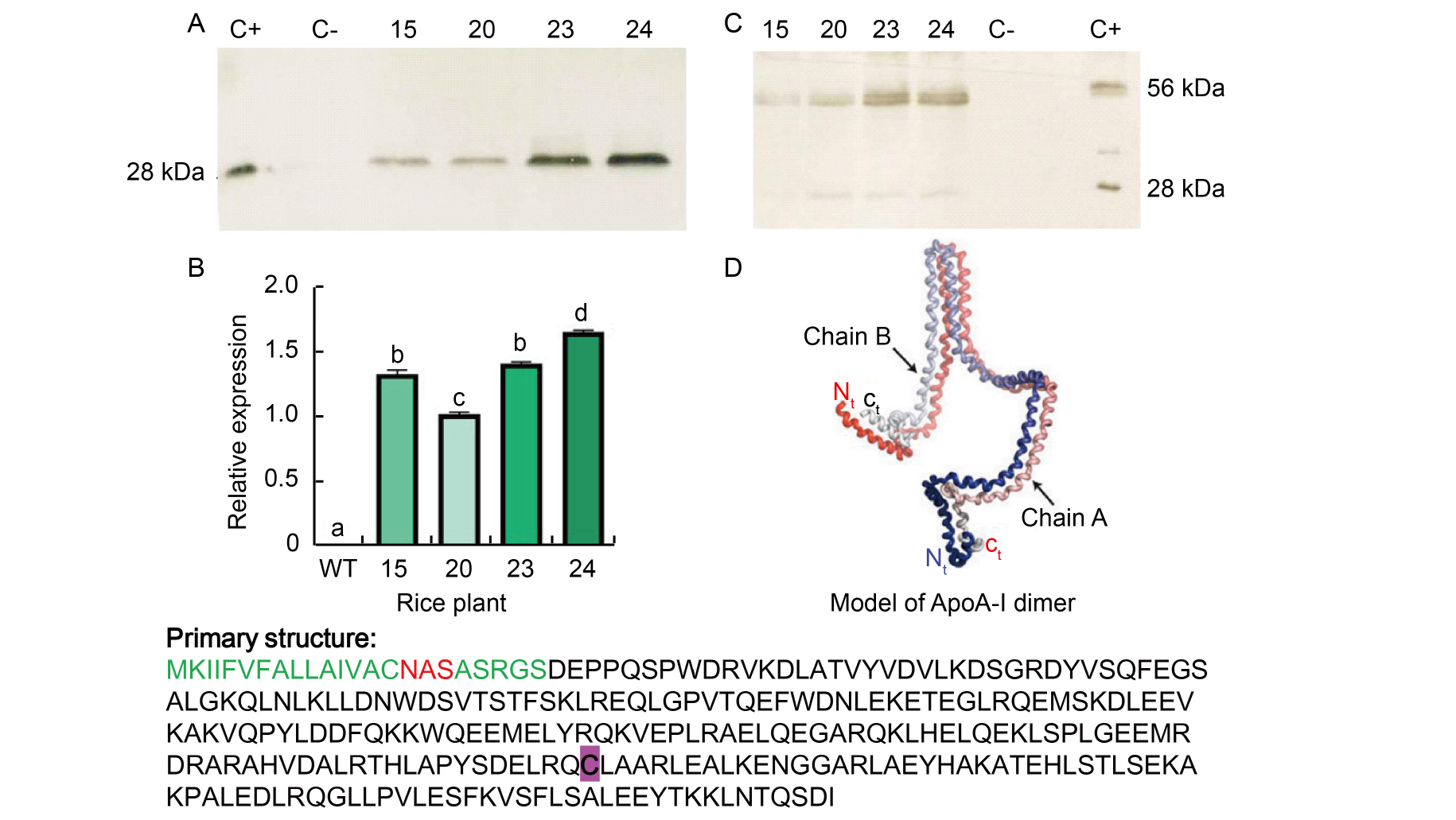
Fig. 1. Western blot under reducing and non-reducing conditions, and the model of ApoA-I dimer by Gogonea et al (2013). A, Western blot under reducing conditions (with β-mercaptoethanol); C+, ApoA-I recombinant; C-, Untransformed rice; ID 15, 20, 23, and 24 rice plants were positive in the screening PCR. All plants showed a band at 28 kDa, similar to the positive control, which was not present in the wild type (WT, untransformed rice) seeds. B, Relative expression of ApoA-IMilano gene in rice plants. RNA was extracted from rice seeds of different plants, and beta-tubulin was used as housekeeping gene. ID 15, 20, 23, and 24 rice plants were positive in the screening PCR. Results are presented as Mean ± SE from three biological replicates. Different lowercase letters above the bars indicate statistically significant differences at the level of P ≤ 0.05.
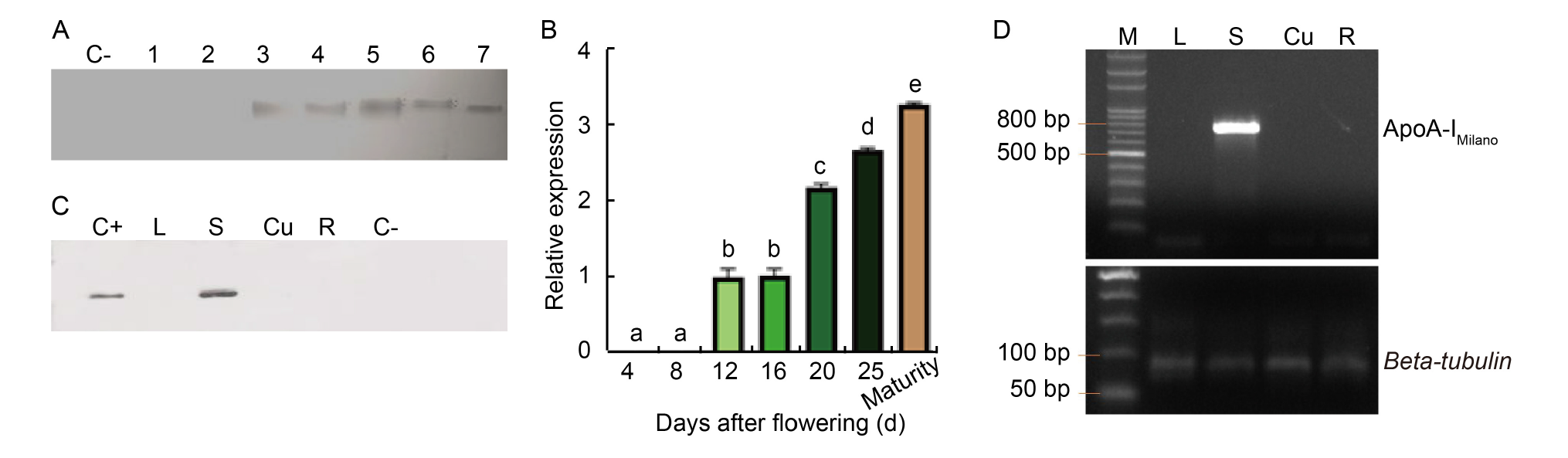
Fig. 2. Temporal and spatial expression of ApoA-IM gene. A, Western blot under reducing conditions (with β-mercaptoethanol). Rice seeds from the same panicle were collected at different days after flowering. Lines 1, 2, 3, 4, 5, 6, and 7 represent 4, 8, 12, 16, 20, and 25 days after flowering, and maturity stage, respectively. B, qRT-PCR analysis of ApoA-IM gene expression levels in seeds at different ripening stages. Results are presented as Mean ± SE from three biological replicates. Different lowercase letters above the bars indicate statistically significant differences at the level of P ≤ 0.05. C, Western blot under reducing conditions was performed on different plant tissues of the same transformed rice plant. The presence of the signal corresponding to positive control confirmed the seed-specificity of 13 kDa promoter. D, RT-PCR carried out on different plant tissues. Beta-tubulin was used as the internal control. C+, Recombinant hApoA-I; L, Proteins extracted from the leaf; S, Proteins extracted from transformed rice seeds; Cu, Proteins extracted from culm; R, Proteins extracted from roots; C-, Rice seeds from the untransformed plant.
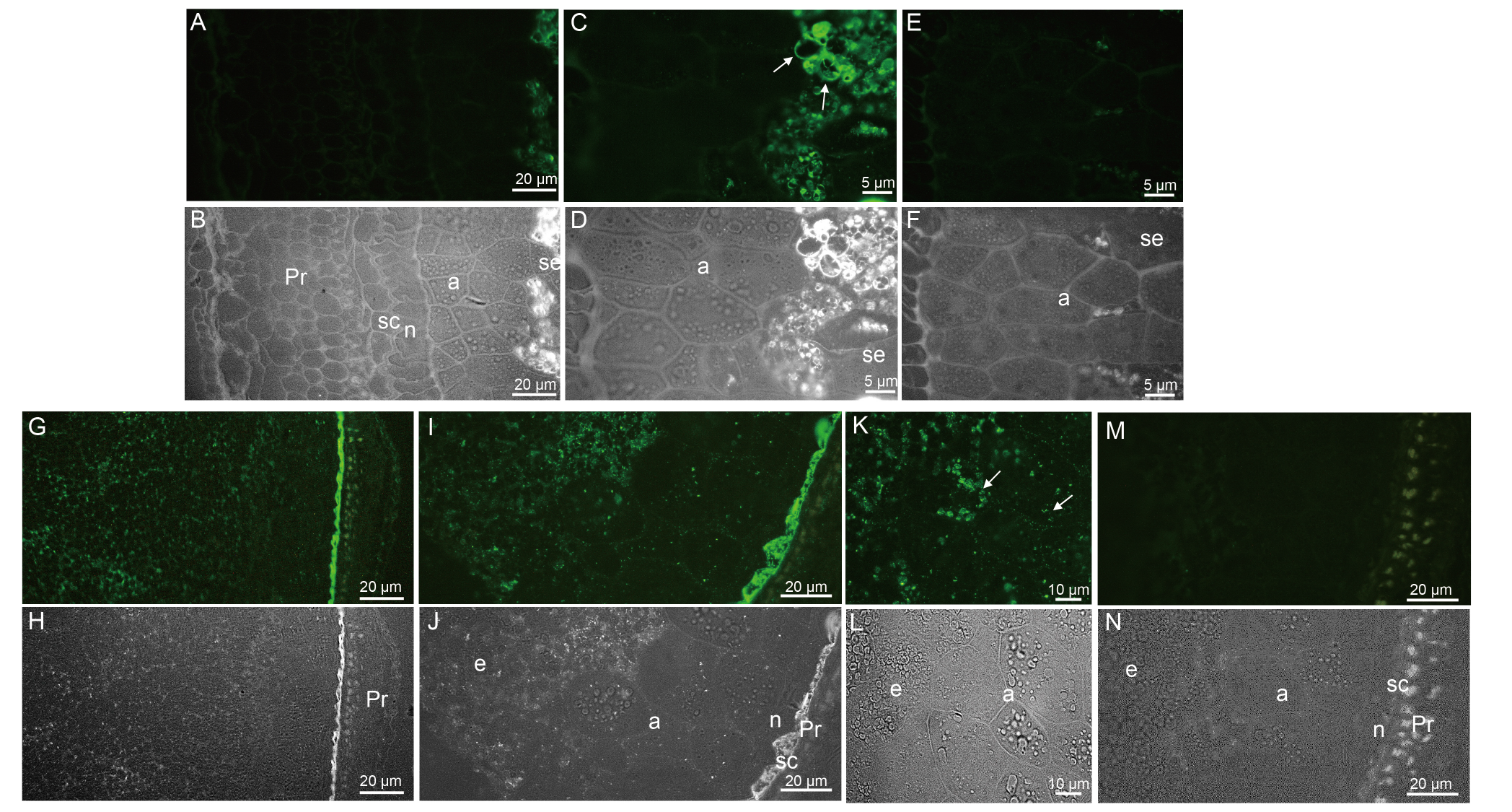
Fig. 3. Immunofluorescence and bright field images of caryopses I (A-F) and II (G-N) of ApoA-IM transformed plants. A-F, Immunofluorescence (A, C, and E) and bright field images (B, D, and F) of caryopsis I of ApoA-IM transformed plant. For A and C, a high fluorescence was observed only in the amyloplasts of endosperm cells particularly in the stroma, surrounding starch granules in C with arrows. E and F, Negative controls showed no cross-reaction with seed tissues. G-N, Immunofluorescence (G, I, K, and M) and bright field images (H, J, L, and N) of caryopsis II of ApoA-IM transformed plant. Pericarp and nucellus appeared thinner in G, H, I, J, M, and N. ApoA-IM was localized in organelles in endosperm cells, seed coats, and aleurone cells in I and K with arrows. G and I, High fluorescence was observed in the seed coat cells. M and N, Only autofluorescence was observed in negative control. a, Aleurons cell; n, Nucellus; Pr, Pericarp; sc, Seed coat; se, Seed endosperm cell.
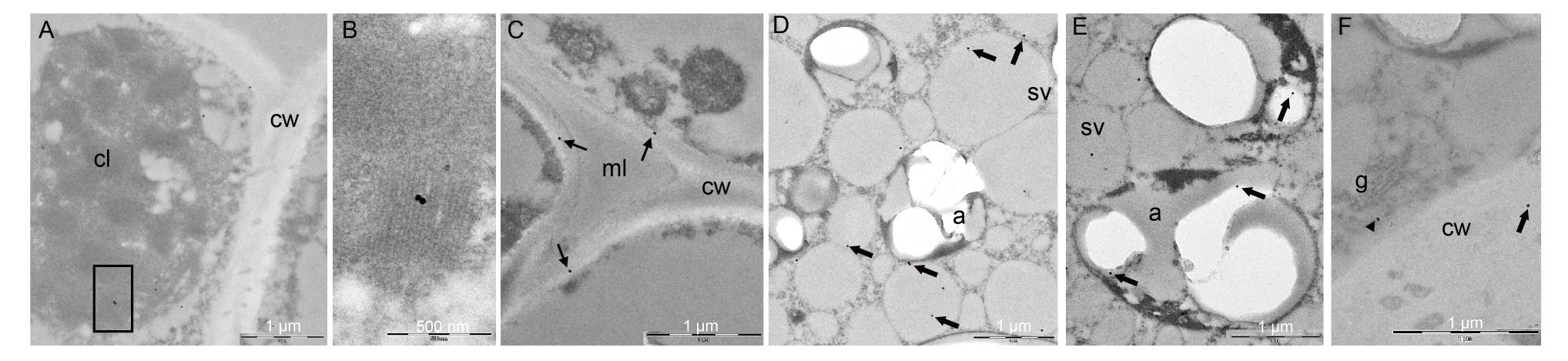
Fig. 4. Immunogold analysis on caryopsis II of ApoA-IM transformed plants. A and B, ApoA-IM was localized into plasmids in the chloroplast grana in the seed coat (B is a magnification of box in A). C, Dark spots were observed in the primary cell wall and not in middle lamellae (with arrows). D, In aleuron cells, ApoA-IM was localized in storage vacuoles (with arrows). E, ApoA-IM was localized into plasmids in amyloplasts of the endosperm cells (with arrows). F, In aleuron cells, ApoA-IM was localized in vesicles associated with dictyiosomes (with arrowhead). cl, Chloroplast; cw, Cell wall; ml, Middle lamellae; a, Amyloplast; sv, Storage vacuole; g, Golgi body.
| [1] | Alexander E T, Tanaka M, Kono M, Saito H, Rader D J, Phillips M C. 2009. Structural and functional consequences of the Milano mutation (R173C) in human apolipoprotein A-I. J Lipid Res, 50(7): 1409-1419. |
| [2] | Baslam M, Oikawa K, Kitajima-Koga A, Kaneko K, Mitsui T. 2016. Golgi-to-plastid trafficking of proteins through secretory pathway: Insights into vesicle-mediated import toward the plastids. Plant Signal Behav, 11(9): e1221558. |
| [3] | Ben-Aicha S, Badimon L, Vilahur G. 2020. Advances in HDL: Much more than lipid transporters. Int J Mol Sci, 21(3): 732. |
| [4] | Chiesa G, Sirtori C R. 2003. Apolipoprotein A-IMilano: Current perspectives. Curr Opin Lipidol, 14(2): 159-163. |
| [5] | Chyu K Y, Shah P K. 2015. HDL/ApoA-1 infusion and ApoA-1 gene therapy in atherosclerosis. Front Pharmacol, 6: 187. |
| [6] | Doyle J J, Doyle J L. 1987. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull, 19(1): 11-15. |
| [7] | Emancipator K, Csako G, Elin R J. 1992. In vitro inactivation of bacterial endotoxin by human lipoproteins and apolipoproteins. Infect Immun, 60(2): 596-601. |
| [8] | Gelvin S B. 2003. Agrobacterium-mediated plant transformation: The biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev, 67(1): 16-37. |
| [9] | Gogonea V, Gerstenecker G S, Wu Z P, Lee X, Topbas C, Wagner M A, Tallant T C, Smith J D, Callow P, Pipich V, Malet H, Schoehn G, DiDonato J A, Hazen S L. 2013. The low-resolution structure of nHDL reconstituted with DMPC with and without cholesterol reveals a mechanism for particle expansion. J Lipid Res, 54(4): 966-983. |
| [10] | Gunasekaran B, Gothandam K M. 2020. A review on edible vaccines and their prospects. Braz J Med Biol Res, 53(2): e8749. |
| [11] | Gupta R, Brunak S. 2002. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac Symp Biocomput, 7: 310-322. |
| [12] | Hiei Y, Ohta S, Komari T, Kumashiro T. 1994. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J, 6(2): 271-282. |
| [13] | Jain N, Vergish S, Khurana J P. 2018. Validation of house-keeping genes for normalization of gene expression data during diurnal/ circadian studies in rice by RT-qPCR. Sci Rep, 8: 3203. |
| [14] | Kim S J, Brandizzi F. 2014. The plant secretory pathway: An essential factory for building the plant cell wall. Plant Cell Physiol, 55(4): 687-693. |
| [15] | Kurup V M, Thomas J. 2020. Edible vaccines: Promises and challenges. Mol Biotechnol, 62(2): 79-90. |
| [16] | Laemmli U K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature, 227: 680-685. |
| [17] | Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods, 25(4): 402-408. |
| [18] | Lorenzetti R, Sidoli A, Palomba R, Monaco L, Martineau D, Lappi D A, Soria M. 1986. Expression of the human apolipoprotein AI gene fused to the E. coligene for β-galactosidase. FEBS Lett, 194(2): 347-350. |
| [19] | Mamat U, Wilke K, Bramhill D, Schromm A B, Lindner B, Kohl T A, Corchero J L, Villaverde A, Schaffer L, Head S R, Souvignier C, Meredith T C, Woodard R W. 2015. Detoxifying Escherichia coli for endotoxin-free production of recombinant proteins. Microb Cell Fact, 14: 57. |
| [20] | Moscatelli A, Gagliardi A, Maneta-Peyret L, Bini L, Stroppa N, Onelli E, Landi C, Scali M, Idilli A I, Moreau P. 2015. Characterisation of detergent-insoluble membranes in pollen tubes of Nicotiana tabacum (L.). Biol Open, 4(3): 378-399. |
| [21] | Muench D G, Ogawa M, Okita T W. 1999. The prolamins of rice. In: Shewry P R, Casey R. Seed Proteins. Dordrecht, the Netherlands: Springer: 93-108. |
| [22] | Nissen S E, Tsunoda T, Tuzcu E M, Schoenhagen P, Cooper C J, Yasin M, Eaton G M, Lauer M A, Sheldon W S, Grines C L, Halpern S, Crowe T, Blankenship J C, Kerensky R. 2003. Effect of recombinant ApoA-I Milano on coronary atherosclerosis in patients with acute coronary syndromes: A randomized controlled trial. JAMA, 290(17): 2292-2300. |
| [23] | Nykiforuk C L, Shen Y, Murray E W, Boothe J G, Busseuil D, Rhéaume E, Tardif J C, Reid A, Moloney M M. 2011. Expression and recovery of biologically active recombinant Apolipoprotein AIMilano from transgenic safflower (Carthamus tinctorius) seeds. Plant Biotechnol J, 9(2): 250-263. |
| [24] | Oluwayelu D O, Adebiyi A I. 2016. Plantibodies in human and animal health: A review. Afr Health Sci, 16(2): 640-645. |
| [25] | Onelli E, Moscatelli A, Gagliardi A, Zaninelli M, Bini L, Baldi A, Caccianiga M, Reggi S, Rossi L. 2017. Retarded germination of Nicotiana tabacum seeds following insertion of exogenous DNA mimics the seed persistent behavior. PLoS One, 12(12): e0187929. |
| [26] | Petrlova J, Dalla-Riva J, Mörgelin M, Lindahl M, Krupinska E, Stenkula K G, Voss J C, Lagerstedt J O. 2014. Secondary structure changes in ApoA-I Milano (R173C) are not accompanied by a decrease in protein stability or solubility. PLoS One, 9(4): e96150. |
| [27] | Qu L Q, Takaiwa F. 2004. Evaluation of tissue specificity and expression strength of rice seed component gene promoters in transgenic rice. Plant Biotechnol J, 2(2): 113-125. |
| [28] | Romano G, Reggi S, Kutryb-Zajac B, Facoetti A, Chisci E, Pettinato M, Giuffrè M R, Vecchio F, Leoni S, De Giorgi M, Avezza F, Cadamuro M, Crippa L, Leone B E, Lavitrano M, Rivolta I, Barisani D, Smolenski R T, Giovannoni R. 2018. APOA-1Milano muteins, orally delivered via genetically modified rice, show anti-atherogenic and anti-inflammatory properties in vitro and in Apoe-/- atherosclerotic mice. Int J Cardiol, 271: 233-239. |
| [29] | Rosano G L, Ceccarelli E A. 2014. Recombinant protein expression in Escherichia coli: Advances and challenges. Front Microbiol, 5: 172. |
| [30] | Rossi L, Dell’Orto V, Vagni S, Sala V, Reggi S, Baldi A. 2014a. Protective effect of oral administration of transgenic tobacco seeds against verocytotoxic Escherichia coli strain in piglets. Vet Res Commun, 38(1): 39-49. |
| [31] | Rossi L, Pinotti L, Agazzi A, Dell’Orto V, Baldi A. 2014b. Plant bioreactors for the antigenic hook-associated flgK protein expression. Ital J Animal Sci, 13(1): 2939. |
| [32] | Ryan R O, Forte T M, Oda M N. 2003. Optimized bacterial expression of human apolipoprotein A-I. Protein Expr Purif, 27(1): 98-103. |
| [33] | Saito Y, Shigemitsu T, Yamasaki R, Sasou A, Goto F, Kishida K, Kuroda M, Tanaka K, Morita S, Satoh S, Masumura T. 2012. Formation mechanism of the internal structure of type I protein bodies in rice endosperm: Relationship between the localization of prolamin species and the expression of individual genes. Plant J, 70(6): 1043-1055. |
| [34] | Schmidt H H J, Genschel J, Haas R, Büttner C, Manns M P. 1997. Expression and purification of recombinant human apolipoprotein A-I in Chinese Hamster ovary cells. Protein Expr Purif, 10(2): 226-236. |
| [35] | Sinha P, Poland J, Schnölzer M, Rabilloud T. 2001. A new silver staining apparatus and procedure for matrix-assisted laser desorption/ionization-time of flight analysis of proteins after two-dimensional electrophoresis. Proteomics, 1(7): 835-840. |
| [36] | Sirtori C R, Ruscica M, Calabresi L, Chiesa G, Giovannoni R, Badimon J J. 2019. HDL therapy today: From atherosclerosis, to stent compatibility to heart failure. Ann Med, 51(7/8): 345-359. |
| [37] | Smith J D. 2010. Apolipoprotein AI and its mimetics for the treatment of atherosclerosis. Curr Opin Invest Drugs, 11(9): 989-996. |
| [38] | Stoffel W. 1984. Synthesis, transport, and processing of apolipoproteins of high density lipoproteins. J Lipid Res, 25(13): 1586-1592. |
| [39] | Streatfield S J. 2007. Approaches to achieve high-level heterologous protein production in plants. Plant Biotechnol J, 5(1): 2-15. |
| [40] | Takaiwa F, Kikuchi S, Oono K. 1987. A rice glutelin gene family: A major type of glutelin mRNAs can be divided into two classes. Mol Gen Genet, 208(1/2): 15-22. |
| [41] | Takaiwa F, Wakasa Y, Hayashi S, Kawakatsu T. 2017. An overview on the strategies to exploit rice endosperm as production platform for biopharmaceuticals. Plant Sci, 263: 201-209. |
| [42] | Tanaka K, Sugimoto T, Ogawa M, Kasai Z. 1980. Isolation and characterization of two types of protein bodies in the rice endosperm. Agric Biol Chem, 44(7): 1633-1639. |
| [43] | Tardif J C, Grégoire J, L’Allier P L, Ibrahim R, Lespérance J, Heinonen T M, Kouz S, Berry C, Basser R, Lavoie M A, Guertin M C, Rodés-Cabau J. 2007. Effects of reconstituted high-density lipoprotein infusions on coronary atherosclerosis: A randomized controlled trial. JAMA, 297: 1675-1682. |
| [44] | Torres E, Gonzalez-Melendi P, Stöger E, Shaw P, Twyman R M, Nicholson L, Vaquero C, Fischer R, Christou P, Perrin Y. 2001. Native and artificial reticuloplasmins co-accumulate in distinct domains of the endoplasmic reticulum and in post-endoplasmic reticulum compartments. Plant Physiol, 127(3): 1212-1223. |
| [45] | Watts D, MacBeath J R. 2001. Automated fluorescent DNA sequencing on the ABI PRISM 310 Genetic Analyzer. Methods Mol Biol, 167: 153-170. |
| [46] | Zhu Q L, Tan J T, Liu Y G. 2022. Molecular farming using transgenic rice endosperm. Trends Biotechnol, 40(10): 1248-1260. |
| [1] | Li Qianlong, Feng Qi, Wang Heqin, Kang Yunhai, Zhang Conghe, Du Ming, Zhang Yunhu, Wang Hui, Chen Jinjie, Han Bin, Fang Yu, Wang Ahong. Genome-Wide Dissection of Quan 9311A Breeding Process and Application Advantages [J]. Rice Science, 2023, 30(6): 552-565. |
| [2] | Ji Dongling, Xiao Wenhui, Sun Zhiwei, Liu Lijun, Gu Junfei, Zhang Hao, Matthew Tom Harrison, Liu Ke, Wang Zhiqin, Wang Weilu. Translocation and Distribution of Carbon-Nitrogen in Relation to Rice Yield and Grain Quality as Affected by High Temperature at Early Panicle Initiation Stage [J]. Rice Science, 2023, 30(6): 598-612. |
| [3] | Prathap V, Suresh Kumar, Nand Lal Meena, Chirag Maheshwari, Monika Dalal, Aruna Tyagi. Phosphorus Starvation Tolerance in Rice Through Combined Physiological, Biochemical, and Proteome Analyses [J]. Rice Science, 2023, 30(6): 613-631. |
| [4] | Sundus Zafar, Xu Jianlong. Recent Advances to Enhance Nutritional Quality of Rice [J]. Rice Science, 2023, 30(6): 523-536. |
| [5] | Kankunlanach Khampuang, Nanthana Chaiwong, Atilla Yazici, Baris Demirer, Ismail Cakmak, Chanakan Prom-U-Thai. Effect of Sulfur Fertilization on Productivity and Grain Zinc Yield of Rice Grown under Low and Adequate Soil Zinc Applications [J]. Rice Science, 2023, 30(6): 632-640. |
| [6] | Fan Fengfeng, Cai Meng, Luo Xiong, Liu Manman, Yuan Huanran, Cheng Mingxing, Ayaz Ahmad, Li Nengwu, Li Shaoqing. Novel QTLs from Wild Rice Oryza longistaminata Confer Strong Tolerance to High Temperature at Seedling Stage [J]. Rice Science, 2023, 30(6): 577-586. |
| [7] | Lin Shaodan, Yao Yue, Li Jiayi, Li Xiaobin, Ma Jie, Weng Haiyong, Cheng Zuxin, Ye Dapeng. Application of UAV-Based Imaging and Deep Learning in Assessment of Rice Blast Resistance [J]. Rice Science, 2023, 30(6): 652-660. |
| [8] | Md. Forshed Dewan, Md. Ahiduzzaman, Md. Nahidul Islam, Habibul Bari Shozib. Potential Benefits of Bioactive Compounds of Traditional Rice Grown in South and Southeast Asia: A Review [J]. Rice Science, 2023, 30(6): 537-551. |
| [9] | Raja Chakraborty, Pratap Kalita, Saikat Sen. Phenolic Profile, Antioxidant, Antihyperlipidemic and Cardiac Risk Preventive Effect of Pigmented Black Rice Variety Chakhao poireiton in High-Fat High-Sugar Induced Rats [J]. Rice Science, 2023, 30(6): 641-651. |
| [10] | Nazaratul Ashifa Abdullah Salim, Norlida Mat Daud, Julieta Griboff, Abdul Rahim Harun. Elemental Assessments in Paddy Soil for Geographical Traceability of Rice from Peninsular Malaysia [J]. Rice Science, 2023, 30(5): 486-498. |
| [11] | Monica Ruffini Castiglione, Stefania Bottega, Carlo Sorce, Carmelina SpanÒ. Effects of Zinc Oxide Particles with Different Sizes on Root Development in Oryza sativa [J]. Rice Science, 2023, 30(5): 449-458. |
| [12] | Tan Jingyi, Zhang Xiaobo, Shang Huihui, Li Panpan, Wang Zhonghao, Liao Xinwei, Xu Xia, Yang Shihua, Gong Junyi, Wu Jianli. ORYZA SATIVA SPOTTED-LEAF 41 (OsSPL41) Negatively Regulates Plant Immunity in Rice [J]. Rice Science, 2023, 30(5): 426-436. |
| [13] | Ammara Latif, Sun Ying, Pu Cuixia, Noman Ali. Rice Curled Its Leaves Either Adaxially or Abaxially to Combat Drought Stress [J]. Rice Science, 2023, 30(5): 405-416. |
| [14] | Liu Qiao, Qiu Linlin, Hua Yangguang, Li Jing, Pang Bo, Zhai Yufeng, Wang Dekai. LHD3 Encoding a J-Domain Protein Controls Heading Date in Rice [J]. Rice Science, 2023, 30(5): 437-448. |
| [15] | Lu Xuedan, Li Fan, Xiao Yunhua, Wang Feng, Zhang Guilian, Deng Huabing, Tang Wenbang. Grain Shape Genes: Shaping the Future of Rice Breeding [J]. Rice Science, 2023, 30(5): 379-404. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||