
Rice Science ›› 2023, Vol. 30 ›› Issue (3): 257-266.DOI: 10.1016/j.rsci.2023.03.009
• Research Paper • Previous Articles
Wei Qinghui1,2,#, Cui Daizong1,#( ), Zheng Baojiang1(
), Zheng Baojiang1( ), Zhao Min1(
), Zhao Min1( )
)
Received:2022-07-01
Accepted:2022-11-09
Online:2023-05-28
Published:2023-03-13
Contact:
Zhao Min (82191513@163.com); Cui Daizong (siyu19831114@163.com); Zheng Baojiang (zbjnefu@126.com)
About author:First author contact:#These authors contributed equally to this work
Wei Qinghui, Cui Daizong, Zheng Baojiang, Zhao Min. In Vitro Antifungal Activity of Dihydrochelerythrine and Proteomic Analysis in Ustilaginoidea virens[J]. Rice Science, 2023, 30(3): 257-266.
Add to citation manager EndNote|Ris|BibTeX
| Pathogen | DHCHE concentration (mg/L) | ||
|---|---|---|---|
| 5.0 | 7.5 | 10.0 | |
| Ustilaginoidea virens | 56.3 ± 1.2 a | 68.8 ± 1.5 b | 75.0 ± 1.0 c |
| Aspergillus ruber | 6.3 ± 1.3 a | 20.3 ± 2.2 b | 30.8 ± 1.7 c |
| Magnaporthe oryzae | 31.7 ± 1.5 a | 44.3 ± 2.0 b | 59.5 ± 1.0 c |
| Nigrospora oryzae | 68.3 ± 2.3 a | 81.3 ± 1.1 b | 84.2 ± 1.2 c |
| Cochliobolus miyabeanus | 36.5 ± 1.0 a | 50.0 ± 1.3 b | 66.2 ± 1.7 c |
Table 1. Growth inhibitory effect of dihydrochelerythrine (DHCHE) on five species of rice pathogenic fungi. %
| Pathogen | DHCHE concentration (mg/L) | ||
|---|---|---|---|
| 5.0 | 7.5 | 10.0 | |
| Ustilaginoidea virens | 56.3 ± 1.2 a | 68.8 ± 1.5 b | 75.0 ± 1.0 c |
| Aspergillus ruber | 6.3 ± 1.3 a | 20.3 ± 2.2 b | 30.8 ± 1.7 c |
| Magnaporthe oryzae | 31.7 ± 1.5 a | 44.3 ± 2.0 b | 59.5 ± 1.0 c |
| Nigrospora oryzae | 68.3 ± 2.3 a | 81.3 ± 1.1 b | 84.2 ± 1.2 c |
| Cochliobolus miyabeanus | 36.5 ± 1.0 a | 50.0 ± 1.3 b | 66.2 ± 1.7 c |
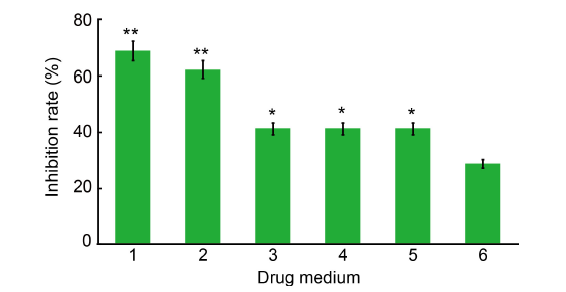
Fig. 1. Inhibition of mycelium growth of Ustilaginoidea virens by dihydrochelerythrine (DHCHE) and other isoquinoline alkaloids. 1, DHCHE; 2, Chelerythrine chloride; 3, Dihydrosanguinarine; 4, Sanguinarine hydrochloride; 5, 6-methoxydihydrosanguinarine; 6, Validamycin (control).The drug media were used at the concentration of 7.5 mg/L. Values are Mean ± SD (n = 3). *, P < 0.05; **, P < 0.01.
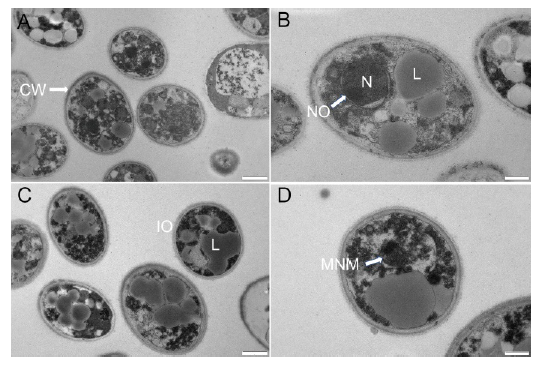
Fig. 2. Transmission electron micrographs of Ustilaginoidea virens spores. A and B, Control group. C and D, Group treated with 15.0 mg/L dihydrochelerythrine. CW, Cell wall; L, Lipid globule; N, Nucleus; NO, Nucleolus; IO, Incomplete organelle; MNM, Malformed nuclear membrane. Scale bars are 1 μm in A and C, and 500 nm in B and D.
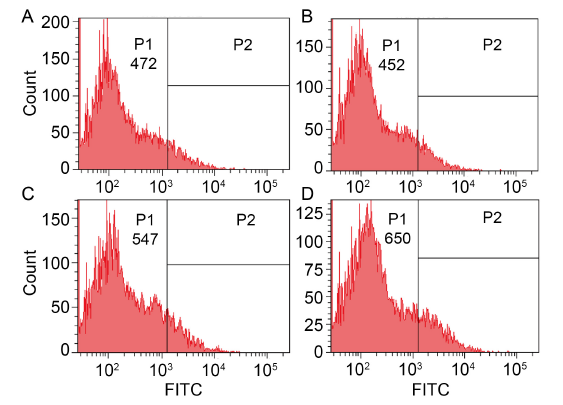
Fig. 3. Effects of dihydrochelerythrine (DHCHE) on reactive oxygen species accumulation. A, Control group; B, 5.0 mg/L DHCHE treatment; C, 7.5 mg/L DHCHE treatment; D, 15.0 mg/L DHCHE treatment. P1 and P2 are groups divided according to the intensity of fluorescence signal, and FITC is fluorescence intensity.
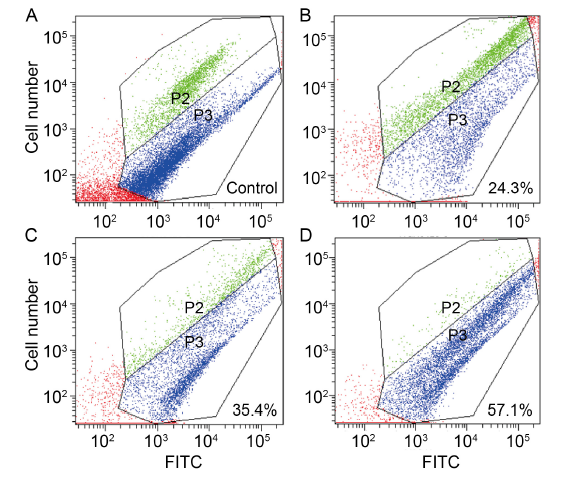
Fig. 4. Effects of dihydrochelerythrine (DHCHE) on mitochondrial membrane potential of Ustilaginoidea virens. A, Positive control; B, 5.0 mg/L DHCHE treatment; C, 7.5 mg/L DHCHE treatment; D, 15.0 mg/L DHCHE treatment. Phylum P2 represents cells with high mitochondrial membrane potential (normal cells), and phylum P3 represents a population with low mitochondrial membrane potential (damaged or apoptotic cells). FITC is fluorescence intensity. With the increase of drug concentration, the proportion of cells in P3 phylum increased significantly, that is, the proportion of cell damage increased.
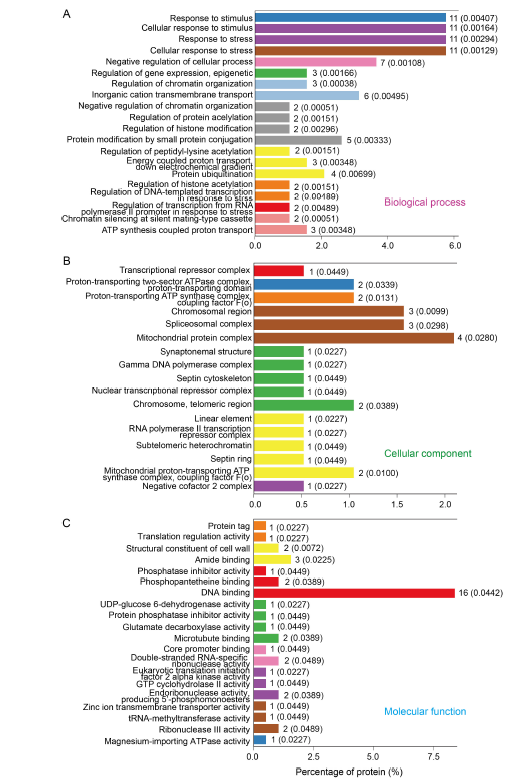
Fig. 5. Gene Ontology (GO) enrichment analysis of differentially expressed proteins in control and dihydrochelerythrine treatment groups (top 20 terms). A?C, Top 20 GO terms in biological process (A), cellular component (B) and molecular function (C).The numbers after the columns and in the parentheses refer to the number of proteins enriched in the corresponding functional level of the GO concentration and the corresponding P values, respectively.
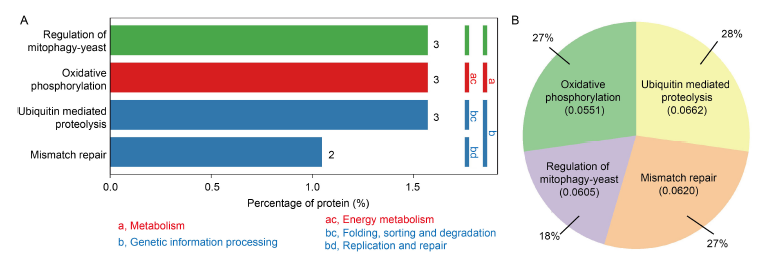
Fig. 6. Enrichment items of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway between control and dihydrochelerythrine treatment groups. A, Differentially expressed proteins (DEPs) in the significantly enriched KEGG pathway. The ordinate on the left shows the specific metabolic pathway enriched. The ordinate on the right shows the abbreviation of the first and second classifications of the metabolic pathway enriched. Green nodes indicate species-specific. Nodes with red boxes indicate up-regulation, while nodes with blue boxes indicate down-regulation. The metabolism is mainly enriched in two different pathways: (1) Metabolism includes one metabolism, namely energy metabolism, and the specific pathway is oxidative phosphorylation; (2) Genetic information processing includes two metabolic pathways, which are folding, sorting and degradation as well as replication and repair. The specific pathway enriched in folding, sorting and degradation is ubiquitin mediated proteolysis, and the replication and repair pathway is concentrated in mismatch repair.B, Pie chart of DEPs. The KEGG enrichment results were sorted according to the P value, and the enriched protein amount in the top 20 items with the smallest P value was counted. Among them, oxidative phosphorylation and mismatch repair accounted for 27%, ubiquitin mediated proteolysis accounted for 28%, and regulation of mitophagy-yeast accounted for 18%.
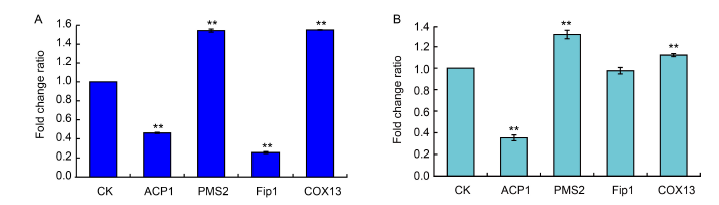
Fig. 7. Average fold change ratios in expression of proteins in Ustilaginoidea virens cells after dihydrochelerythrine (DHCHE) treatment. A, Protein expression in DHCHE-treated U. virens cells by tandem mass tag (TMT)-based quantitative proteomics assay. B, Protein expression in DHCHE-treated U. virens cells by parallel reaction monitoring (PRM) assay. U. virens in the treatment groups was cultured in potato sucrose medium at 28 oC for 10 d, followed by incubation with 7.5 mg/L of DHCHE for 24 h. No DHCHE was added to the control (CK) groups. Values are Mean ± SD (n = 3). **, P < 0.01.
| [1] | An P P, Yang X B, Yu J, Qi J R, Ren X Y, Kong Q J. 2019. α-terpineol and terpene-4-ol, the critical components of tea tree oil, exert antifungal activities in vitro and in vivo against Aspergillus niger in grapes by inducing morphous damage and metabolic changes of fungus. Food Control, 98: 42-53. |
| [2] |
Branen J K, Chiou T J, Engeseth N J. 2001. Overexpression of acyl carrier protein-1 alters fatty acid composition of leaf tissue in Arabidopsis. Plant Physiol, 127(1): 222-229.
PMID |
| [3] | Cao F J, Yang R, Lv C, Ma Q, Lei M, Geng H L, Zhou L. 2015. Pseudocyanides of sanguinarine and chelerythrine and their series of structurally simple analogues as new anticancer lead compounds: Cytotoxic activity, structure-activity relationship and apoptosis induction. Eur J Pharm Sci, 67: 45-54. |
| [4] | Chen Q L, Tang H L, Zha Z Q, Yin H P, Wang Y, Wang Y F, Li H T, Yue L. 2017. β-d-glucan from Antrodia camphorata ameliorates LPS-induced inflammation and ROS production in human hepatocytes. Int J Biol Macromol, 104: 768-777. |
| [5] | Egan J M, Kaur A, Raja H A, Kellogg J J, Oberlies N H, Cech N B. 2016. Antimicrobial fungal endophytes from the botanical medicine goldenseal (Hydrastis canadensis). Phytochem Lett, 17: 219-225. |
| [6] |
Forbes K P, Addepalli B, Hunt A G. 2006. An Arabidopsis Fip1 homolog interacts with RNA and provides conceptual links with a number of other polyadenylation factor subunits. J Biol Chem, 281(1): 176-186.
PMID |
| [7] | Ghorbanpour M, Omidvari M, Abbaszadeh-Dahaji P, Omidvar R, Kariman K. 2018. Mechanisms underlying the protective effects of beneficial fungi against plant diseases. Biol Control, 117: 147-157. |
| [8] | Guo W W, Gao Y X, Yu Z M, Xiao Y H, Zhang Z G, Zhang H F. 2019. The adenylate cyclase UvAc1 and phosphodiesterase UvPdeH control the intracellular cAMP level, development, and pathogenicity of the rice false smut fungus Ustilaginoidea virens. Fungal Genet Biol, 129: 65-73. |
| [9] |
Helmling S, Zhelkovsky A, Moore C L. 2001. Fip1 regulates the activity of poly(A) polymerase through multiple interactions. Mol Cell Biol, 21(6): 2026-2037.
PMID |
| [10] | Hou F, Cui M H. 2014. Expression and clinical significance of hMLH1 and hPMS2 in colon cancer and concomitant polyps. Chin J Curr Adv Gen Surg, 17(7): 531-534. (in Chinese with English abstract) |
| [11] | Jang B C, Park J G, Song D K, Baek W K, Yoo S K, Jung K H, Park G Y, Lee T Y, Suh S I. 2009. Sanguinarine induces apoptosis in A549 human lung cancer cells primarily via cellular glutathione depletion. Toxicol Vitro, 23(2): 281-287. |
| [12] |
Lauridsen P E, Rasmussen L J, Desler C. 2019. Mitochondrial oxidative phosphorylation capacity of cryopreserved cells. Mitochondrion, 47: 47-53.
PMID |
| [13] | Liu G X. 2012. Effects on lipid synthesis of ACP1 and ACP2 genes on Chlamydomonas reinhardtii. Wuhan, China: Hubei University. (in Chinese with English abstract) |
| [14] | Liu Y J, Li X J, Yuan H, Liu X, Gao Y X, Gong M, Zhou Z R. 2017. Fusing the acyl carrier protein enhances the solubility and thermostability of the recombinant proteins in Escherichia coli. China Biotechnol, 37(7): 115-123. (in Chinese with English abstract) |
| [15] | Lu W. 2018. Mitochondrial oxidative phosphorylation deficiency inhibits the differentiation of brown adipocytes and related mechanism. Suzhou, China: Soochow University. (in Chinese with English abstract) |
| [16] |
Marampon F, Gravina G L, Ju X M, Vetuschi A, Sferra R, Casimiro M C, Pompili S, Festuccia C, Colapietro A, Gaudio E, di Cesare E, Tombolini V, Pestell R G. 2016. Cyclin D1 silencing suppresses tumorigenicity, impairs DNA double strand break repair and thus radiosensitizes androgen-independent prostate cancer cells to DNA damage. Oncotarget, 7: 64526.
PMID |
| [17] | Navarro V, Delgado G. 1999. Two antimicrobial alkaloids from Bocconia arborea. J Ethnopharmacol, 66(2): 223-226. |
| [18] |
Norris A M, Woodruff R D, D’Agostino R B, Clodfelter J E, Scarpinato K D. 2007. Elevated levels of the mismatch repair protein PMS2 are associated with prostate cancer. Prostate, 67(2): 214-225.
PMID |
| [19] |
Perrone G G, Tan S X, Dawes I W. 2008. Reactive oxygen species and yeast apoptosis. Biochim Biophys Acta, 1783(7): 1354-1368.
PMID |
| [20] | Sun J, Zhou J, Wang Z H, He W N, Zhang D J, Tong Q Q, Su X R. 2015. Multi-omics based changes in response to cadmium toxicity in Bacillus licheniformis A. RSC Adv, 5(10): 7330-7339. |
| [21] | Téllez-Robledo B, Manzano C, Saez A, Navarro-Neila S, Silva- Navas J, de Lorenzo L, González-García M P, Toribio R, Hunt A G, Baigorri R, Casimiro I, Brady S M, Castellano M M,Del Pozo J C. 2019. The polyadenylation factor FIP1 is important for plant development and root responses to abiotic stresses. Plant J, 99(6): 1203-1219. |
| [22] | Thomas J. 2009. Characterization of the Escherichia coli acyl carrier protein phosphodiesterase. Illinois, USA: University of Illinois at Urbana-Champaign. |
| [23] | Vrba J, Dolezel P, Vicar J, Modrianský M, Ulrichová J. 2008. Chelerythrine and dihydrochelerythrine induce G1 phase arrest and bimodal cell death in human leukemia HL-60 cells. Toxicol Vitro, 22(4): 1008-1017. |
| [24] |
Wei Q H, Zhao M, Li X Y. 2017. Extraction of chelerythrine and its effects on pathogenic fungus spore germination. Pharmacogn Mag, 13(52): 600-606.
PMID |
| [25] | Wen S Q, Jeyakkumar P, Avula S R, Zhang L, Zhou C H. 2016. Discovery of novel berberine imidazoles as safe antimicrobial agents by down regulating ROS generation. Bioorg Med Chem Lett, 26(12): 2768-2773. |
| [26] | Xin W J, Mao Y S, Lu F, Li T, Wang J X, Duan Y B, Zhou M G. 2020. In vitro fungicidal activity and in planta control efficacy of coumoxystrobin against Magnaporthe oryzae. Pestic Biochem Physiol, 162: 78-85. |
| [27] | Yakovlev A Y, Borovskii G B, Voinikov V K, Grabelnych O I, Pobezhimova T P, Antipina A I. 2002. An effect of water-soluble proteins of fungi differed in frost-hardy on the energetic activity of isolated plant mitochondria. J Therm Biol, 27(3): 239-244. |
| [28] | Yang R, Gao Z F, Zhao J Y, Li W B, Zhou L, Miao F. 2015. New class of 2-aryl-6-chloro-3,4-dihydroisoquinolinium salts as potential antifungal agents for plant protection: Synthesis, bioactivity and structure-activity relationships. J Agric Food Chem, 63(7): 1906-1914. |
| [29] | Yuan J, Guan L J, Cong B, Zhang Z J, Wang L Z. 2005. Determination of antimicrobial activity of matrine alkaloids. J Pestic, 44(2): 4. (in Chinese with English abstract) |
| [30] | Zhao Z M, Shang X F, Lawoe R K, Liu Y Q, Zhou R, Sun Y, Yan Y F, Li J C, Yang G Z, Yang C J. 2019. Anti-phytopathogenic activity and the possible mechanisms of action of isoquinoline alkaloid sanguinarine. Pestic Biochem Physiol, 159: 51-58. |
| [1] | Zhang Guomei, Li Han, Liu Shanshan, Zhou Xuming, Lu Mingyang, Tang Liang, Sun Lihua. Water Extract of Rice False Smut Balls Activates Nrf2/HO-1 and Apoptosis Pathways, Causing Liver Injury [J]. Rice Science, 2023, 30(5): 473-485. |
| [2] | Liu Yueran, Qu Jinsong, Wang Yufu, Yin Weixiao, Luo Chaoxi. bZIP Transcription Factor UvATF21 Mediates Vegetative Growth, Conidiation, Stress Tolerance and Is Required for Full Virulence of Rice False Smut Fungus Ustilaginoidea virens [J]. Rice Science, 2023, 30(1): 50-57. |
| [3] | Meng Shuai, Qiu Jiehua, Xiong Meng, Liu Zhiquan, Jane Sadhna Jagernath, Lin Fucheng, Shi Huanbin, Kou Yanjun. UvWhi2 Is Required for Stress Response and Pathogenicity in Ustilaginoidea virens [J]. Rice Science, 2022, 29(1): 47-54. |
| [4] | Tianqiao Song, Xiong Zhang, You Zhang, Dong Liang, Jiaoling Yan, Junjie Yu, Mina Yu, Huijuan Cao, Mingli Yong, Xiayan Pan, Zhongqiang Qi, Yan Du, Rongsheng Zhang, Yongfeng Liu. Genome-Wide Identification of Zn2Cys6 Class Fungal-Specific Transcription Factors (ZnFTFs) and Functional Analysis of UvZnFTF1 in Ustilaginoidea virens [J]. Rice Science, 2021, 28(6): 567-578. |
| [5] | Junjie Yu, Mina Yu, Tianqiao Song, Huijuan Cao, Mingli Yong, Xiayan Pan, Zhongqiang Qi, Yan Du, Rongsheng Zhang, Xiaole Yin, Dong Liang, Yongfeng Liu. UvSMEK1, a Suppressor of MEK Null, Regulates Pathogenicity, Conidiation and Conidial Germination in Rice False Smut Fungus Ustilaginoidea virens [J]. Rice Science, 2021, 28(5): 457-465. |
| [6] | Meng Xiong, Shuai Meng, Jiehua Qiu, Huanbin Shi, Xiangling Shen, Yanjun Kou. Putative Phosphatase UvPsr1 Is Required for Mycelial Growth, Conidiation, Stress Response and Pathogenicity in Ustilaginonidea virens [J]. Rice Science, 2020, 27(6): 529-536. |
| [7] | Jiehua Qiu, Shuai Meng, Yizhen Deng, Shiwen Huang, Yanjun Kou. Ustilaginoidea virens: A Fungus Infects Rice Flower and Threats World Rice Production [J]. Rice Science, 2019, 26(4): 199-206. |
| Viewed | ||||||
|
Full text |
|
|||||
|
Abstract |
|
|||||