
Rice Science ›› 2020, Vol. 27 ›› Issue (2): 133-142.DOI: 10.1016/j.rsci.2020.01.004
收稿日期:2018-12-16
接受日期:2019-04-15
出版日期:2020-03-28
发布日期:2019-11-28
. [J]. Rice Science, 2020, 27(2): 133-142.
| Gene | Primer (5´ to 3´ ) |
|---|---|
| For vector construction cOsSRK F cOsSRK R gOsSRK F gOsSRK R For RT-PCR rOsSRK F rOsSRK R rUBQ F rUBQ R For Real Time-PCR OsMyb4 F OsMyb4 R OsDREB1A F OsDREB1A R ZOS3-22 F ZOS3-22 R OsWRKY8 F OsWRKY8 R EL5 F EL5 R OsKRP1F OsKRP1 R OsKRP4 F OsKRP4 R OsCDKB2-1 F OsCDKB2-1 R OsCYCA3-1 F OsCYCA3-1 R OsCYCD2-1 F OsCYCD2-1 R OsCYCD5-1 F OsCYCD5-1 R qUBQ F qUBQ R | ATGGATCCCACCAACATGTCTG GCACTAGTACCTGCTCTTAAC ATTCTAGATGTCTGCTAATCCCAAC ATGGATCCAACAATGTAATGATCCAATTG CAACAAGCAAGCCTCCAGCAA CGAAGCAGAAGCCGAACAGC ATCACGCTGGAGGTGGAG AGGCCTTCTGGTTGTAGACG ACAACACCACGGACAGTTT GTGCTGTCCACCGTCATT TACCACACTCGAGCAGAGCA GACGACTCGCCGCTCATCT CGTTCCCGTCGTTCCAG GCTTCTCCTGCACCTTCTT GAACTAGGCCGAAACCCTAAA TTCGTCTTCTTCAGCGACTG TCGGTAGAACCGAACTGGAA GATGCTTCTCACTGCCATCC CAGCCGTCCACTACCTTCTG ATCTGCACGCTCGCTTCAGC CGATTAGCACCCCTGGATCT TCAGTCTAGCTTGACCCATTC CATGGACACCGACCTCAAGA TGCAGAGCTGGTACATCAGG TGGGCAATCCTACAACCAAG TGAACTCCAAGGGAAGGCTA CGAGCATCATCATAACAAGC CATCCTCACATCTCCAATCT ACAACAGGAGGAGCAGCAAG CGGTGTCGGTAACATCATCA ACCACTTCGACCGCCACTACT ACGCCTAAGCCTGCTGGTT |
Supplemental Table 1. Primers used in this study.
| Gene | Primer (5´ to 3´ ) |
|---|---|
| For vector construction cOsSRK F cOsSRK R gOsSRK F gOsSRK R For RT-PCR rOsSRK F rOsSRK R rUBQ F rUBQ R For Real Time-PCR OsMyb4 F OsMyb4 R OsDREB1A F OsDREB1A R ZOS3-22 F ZOS3-22 R OsWRKY8 F OsWRKY8 R EL5 F EL5 R OsKRP1F OsKRP1 R OsKRP4 F OsKRP4 R OsCDKB2-1 F OsCDKB2-1 R OsCYCA3-1 F OsCYCA3-1 R OsCYCD2-1 F OsCYCD2-1 R OsCYCD5-1 F OsCYCD5-1 R qUBQ F qUBQ R | ATGGATCCCACCAACATGTCTG GCACTAGTACCTGCTCTTAAC ATTCTAGATGTCTGCTAATCCCAAC ATGGATCCAACAATGTAATGATCCAATTG CAACAAGCAAGCCTCCAGCAA CGAAGCAGAAGCCGAACAGC ATCACGCTGGAGGTGGAG AGGCCTTCTGGTTGTAGACG ACAACACCACGGACAGTTT GTGCTGTCCACCGTCATT TACCACACTCGAGCAGAGCA GACGACTCGCCGCTCATCT CGTTCCCGTCGTTCCAG GCTTCTCCTGCACCTTCTT GAACTAGGCCGAAACCCTAAA TTCGTCTTCTTCAGCGACTG TCGGTAGAACCGAACTGGAA GATGCTTCTCACTGCCATCC CAGCCGTCCACTACCTTCTG ATCTGCACGCTCGCTTCAGC CGATTAGCACCCCTGGATCT TCAGTCTAGCTTGACCCATTC CATGGACACCGACCTCAAGA TGCAGAGCTGGTACATCAGG TGGGCAATCCTACAACCAAG TGAACTCCAAGGGAAGGCTA CGAGCATCATCATAACAAGC CATCCTCACATCTCCAATCT ACAACAGGAGGAGCAGCAAG CGGTGTCGGTAACATCATCA ACCACTTCGACCGCCACTACT ACGCCTAAGCCTGCTGGTT |
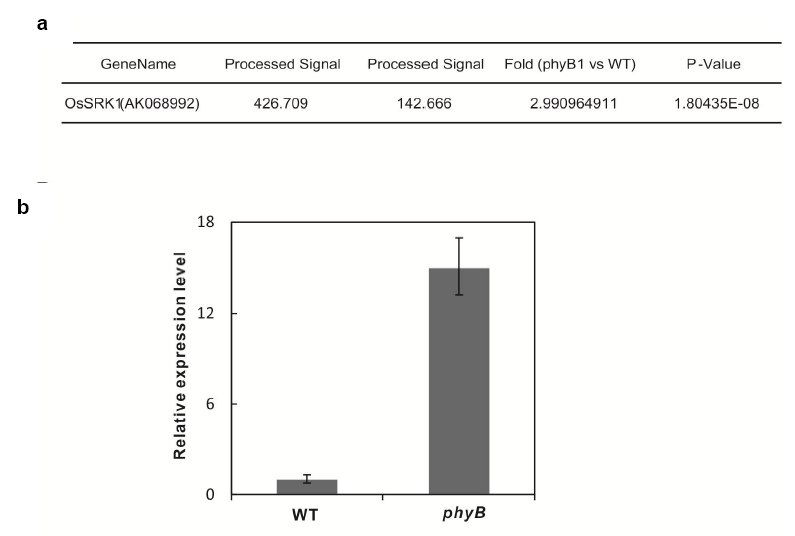
Supplemental Fig. 1. The expression level of OsSRK1 in wild type (WT) and phyB1. a. The expression level of OsSRK1 in WT and phyB1 from genechip results. b. The expression level of OsSRK1 tested by qPCR. Data are means and SE of three biological replicates.

Supplemental Fig. 2. Identity between OsSRK1 and the four reported SRK proteins based on amino acid sequence of serine/threonine-protein kinase domain.
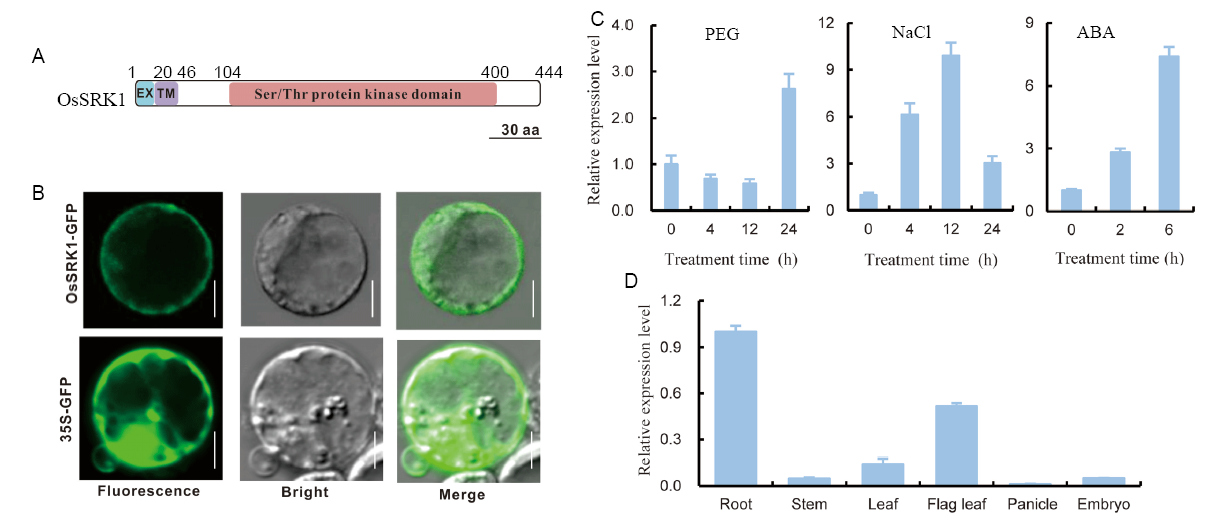
Fig. 1. Subcellular localization and gene expression pattern analysis. A, Schematic representation of OsSRK1. EX, External; TM, Transmembrane. B, Subcellular localization of OsSRK1 protein. Bars = 10 µm. C, Relative expression levels of OsSRK1 in rice leaves treated with 15% polyethylene glycol 4000 (PEG), 150 mmol/L NaCl and 100 μmol/L abscisic acid (ABA). D, Tissue expression pattern of OsSRK1. Data are Mean ± SE (n = 3).
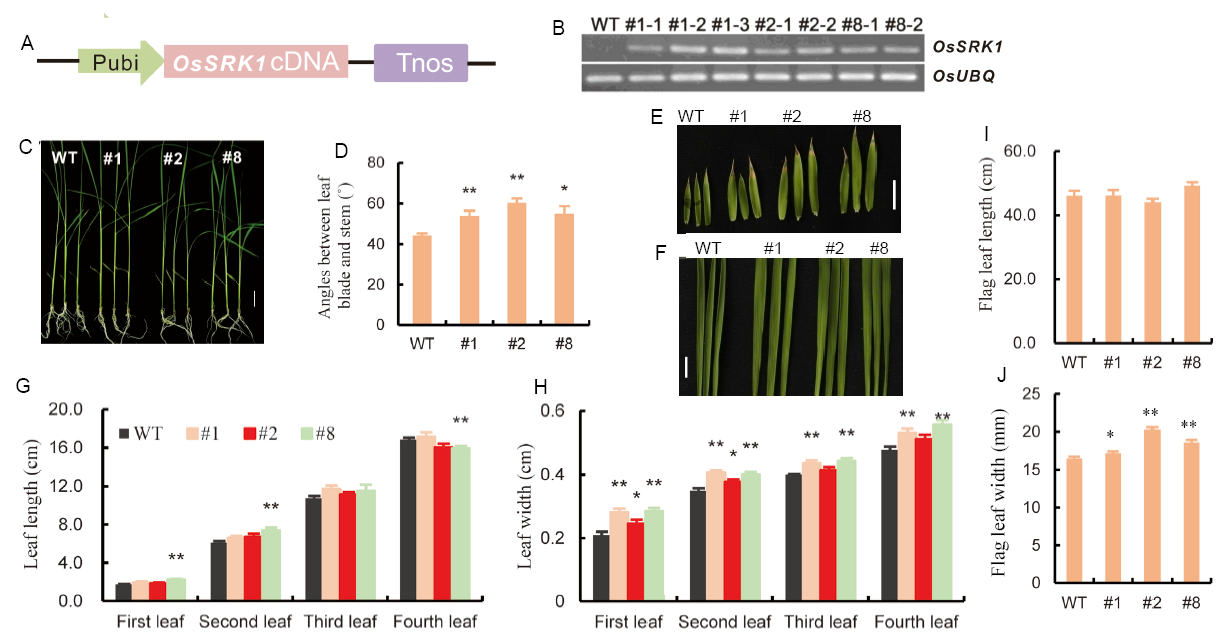
Fig. 2. Overexpression of OsSRK1 affected leaf development. A, Schematic diagrams of the construct used to generate transgenic OsSRK1-overexpression (OsSRK1-OX) rice lines. The expression of OsSRK1 was driven by the ubiquitin promoter. B, Analysis of OsSRK1 expression in transgenic and wild type (WT) plants. #1-1, #1-2 and #1-3 represent three individual plants of OsSRK1-OX line #1; #2-1 and #2-2 represent two individual plants of OsSRK1-OX line #2; #8-1 and #8-2 represent two individual plants of OsSRK1-OX line #8. C, Phenotypes of transgenic seedlings at the 4-leaf stage. Bar = 1 cm. D, Leaf angle of WT and OsSRK1-OX plants. E, Phenotypes of the second leaves. Bar = 1 cm. F, Phenotypes of the third leaves. Bar = 1 cm. G, Leaf length of the first, second, third and fourth leaves. H, Leaf width of the first, second, third and fourth leaves. I, Flag leaf length. J, Flag leaf width. #1, #2 and #8 represent three independent transgenic OsSRK1-OX lines. Data are Mean ± SE (n = 12). Significant differences were determined with the Student’s t-test (*, P < 0.05; **, P < 0.01).
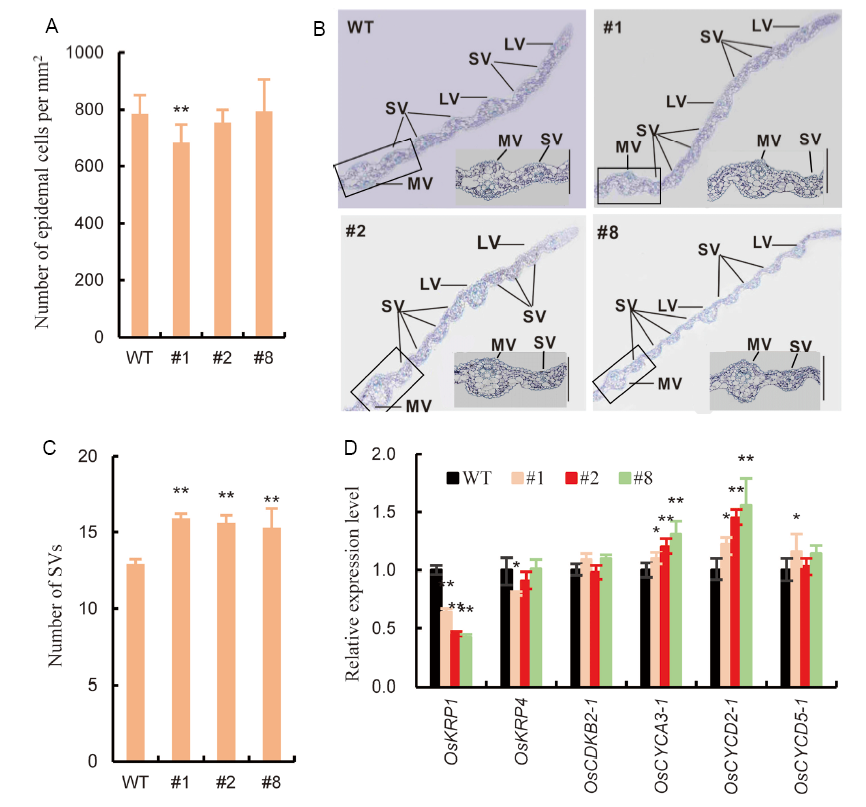
Fig. 3. Overexpression of OsSRK1 affected cell division in leaf. A, Number of non-stomata epidermal cells per mm2 in the fourth leaves of WT and OsSRK1-OX lines. Values are Mean ± SE (n = 5). B, Transverse sections of leaves in WT and OsSRK1-OX lines. MV, Middle vascular bundle; LV, Large vascular bundle; SV, Small vascular bundle. Insets are enlargements showing image of MV and SV. C, Number of small vascular bundles in WT and OsSRK1-OX lines. Values are Mean ± SE (n = 15 to 20). D, Transcript levels of cell division-related genes in the leaf primordium of OsSRK1-OX lines and WT. Values are Mean ± SE (n = 3).WT, Wild type. #1, #2 and #8 represent three independent transgenic OsSRK1-over-expression lines. Significant differences were determined with the Student’s t-test (*, P < 0.05; **, P < 0.01).
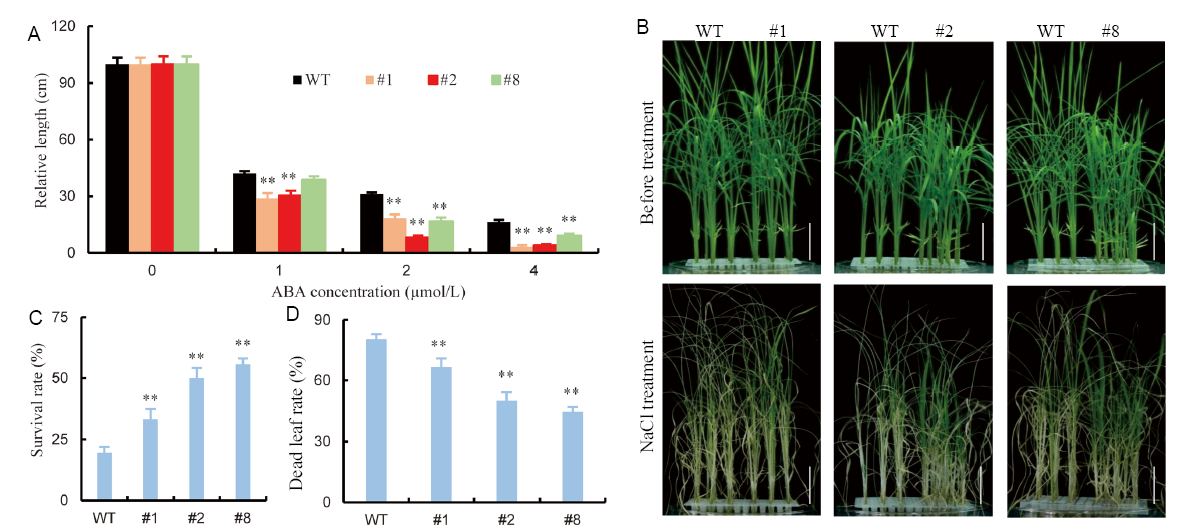
Fig. 4. Overexpression of OsSRK1 enhanced abscisic acid (ABA) sensitivity and salt tolerance. A, Plant heights of OsSRK1-OX (#1, #2 and #3) and wild type (WT) plants treated with exogenous ABA. Values are Mean ± SE (n = 30). B, Tolerance assays of OsSRK1-OX lines to NaCl. Bar = 1 cm. C, Survival rates of WT and the OsSRK1-OX lines treated with 150 mmol/L NaCl for 4 d. Values are Mean ± SE (n = 15 to 20). D, Dead leaf rate of WT and the OsSRK1-OX lines treated with 150 mmol/L NaCl for 4 d. Values are Mean ± SE (n = 15 to 20). Significant differences were determined with the Student’s t-test (*, P < 0.05; **, P < 0.01).
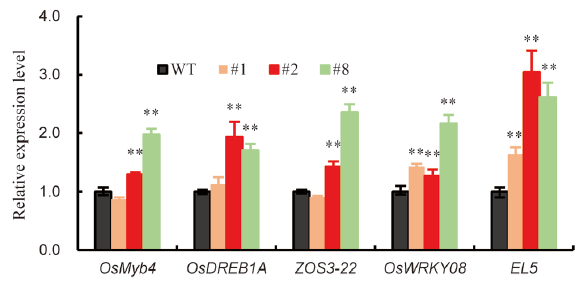
Fig. 5. Transcript levels of genes upregulated in the OsSRK1- overexpressing lines. Three OsSRK1-OX lines (#1, #2 and #8) and wild type (WT) plants were cultured for 14 d in a growth chamber under 14 h light (28 °C)/10 h dark (25 °C). Aboveground parts were harvested to analyze the transcript levels of genes up-regulated by OsSRK1 in the microarray results. Values are Mean ± SE (n = 3). Significant differences were determined with Student’s t-test (*, P < 0.05; **, P < 0.01).
| [1] | Becraft P W. 2002. Receptor kinase signaling in plant development. Annu Rev Cell Dev Biol, 18: 163-192. |
| [2] | Chen L J, Wuriyanghan H, Zhang Y Q, Duan K X, Chen H W, Li Q T, Lu X, He S J, Ma B, Zhang W K, Lin Q, Chen S Y, Zhang J S. 2013. An S-domain receptor-like kinase, OsSIK2, confers abiotic stress tolerance and delays dark-induced leaf senescence in rice. Plant Physiol, 163(4): 1752-1765. |
| [3] | Chen X W, Shang J J, Chen D X, Lei C L, Zou Y, Zhai W X, Liu G Z, Xu J C, Ling Z Z, Cao G, Ma B T, Wang Y P, Zhao X F, Li S G, Zhu L H. 2006. A B-lectin receptor kinase gene conferring rice blast resistance. Plant J, 46(5): 794-804. |
| [4] | Clark S E, Williams R W, Meyerowitz E M. 1997. The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell, 89(4): 575-585. |
| [5] | de Veylder L, Beeckman T, Beemster G T S, Krols L, Terras F, Landrieu I, van der Schueren E, Maes S, Naudts M, Inzé D. 2001. Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. Plant Cell, 13(7): 1653-1668. |
| [6] | Dewitte W, Murray J A H. 2003. The plant cell cycle. Annu Rev Plant Biol, 54: 235-264. |
| [7] | Dubouzet J G, Sakuma Y, Ito Y, Kasuga M, Dubouzet E G, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. 2003. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold- responsive gene expression. Plant J, 33(4): 751-763. |
| [8] | Fan J B, Bai P F, Ning Y S, Wang J Y, Shi X T, Xiong Y H, Zhang K, He F, Zhang C Y, Wang R Y, Meng X Z, Zhou J G, Wang M, Shirsekar G, Park C H, Bellizzi M, Liu W D, Jeon J S, Xia Y, Shan L B, Wang G L. 2018. The monocot-specific receptor-like kinase SDS2 controls cell death and immunity in rice. Cell Host Microbe, 23(4): 498-510. |
| [9] | Gómez-Gómez L, Boller T. 2000. FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell, 5(6): 1003-1011. |
| [10] | Hiei Y, Ohta S, Komari T, Kumashiro T. 1994. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J, 6(2): 271-282. |
| [11] | Irizarry R A, Hobbs B, Collin F, Beazer-Barclay Y D, Antonellis K J, Scherf U, Speed T P. 2003. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics, 4(2): 249-264. |
| [12] | Kinoshita T, Caño-Delgado A, Seto H, Hiranuma S, Fujioka S, Yoshida S, Chory J. 2005. Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature, 433: 167-171. |
| [13] | Livak K J, Schmittgen T D. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods, 25(4): 402-408. |
| [14] | Morgan D O. 1995. Principles of CDK regulation. Nature, 374: 131-134. |
| [15] | Nasrallah J B, Nasrallah M E. 2014. S-locus receptor kinase signalling. Biochem Soc Trans, 42: 313-319. |
| [16] | Ouyang S Q, Liu Y F, Liu P, Lei G, He S J, Ma B, Zhang W K, Zhang J S, Chen S Y. 2010. Receptor-like kinase OsSIK1 improves drought and salt stress tolerance in rice (Oryza sativa) plants. Plant J, 62(2): 316-329. |
| [17] | Park M R, Yun K Y, Mohanty B, Herath V, Xu F Y, Wijaya E, Bajic V B, Yun S J, de Los Reyes B G.2010. Supra-optimal expression of the cold-regulated OsMyb4 transcription factor in transgenic rice changes the complexity of transcriptional network with major effects on stress tolerance and panicle development. Plant Cell Environ, 33: 2209-2230. |
| [18] | Pasquali G, Biricolti S, Locatelli F, Baldoni E, Mattana M. 2008. Osmyb4 expression improves adaptive responses to drought and cold stress in transgenic apples. Plant Cell Rep, 27(10): 1677-1686. |
| [19] | Sherr C J, Roberts J M. 1999. CDK inhibitors: Positive and negative regulators of G1-phase progression. Gene Dev, 13: 1501-1512. |
| [20] | Shiu S H, Bleecker A B. 2001. Plant receptor-like kinase gene family: Diversity, function, and signaling. Sci STKE, 2001: re22. |
| [21] | Soltész A, Vágújfalvi A, Rizza F, Kerepesi I, Galiba G, Cattivelli L, Coraggio I, Crosatti C. 2012. The rice Osmyb4 gene enhances tolerance to frost and improves germination under unfavourable conditions in transgenic barley plants. J Appl Genet, 53(2): 133-143. |
| [22] | Song Y, Jing S J, Yu D Q. 2009. Overexpression of the stress- induced OsWRKY08 improves osmotic stress tolerance in Arabidopsis. Chin Sci Bull, 54: 4671-4678. |
| [23] | Sun M Z, Qian X, Chen C, Cheng S F, Jia B W, Zhu Y M, Sun X L. 2018. Ectopic expression of GsSRK in Medicago sativa reveals its involvement in plant architecture and salt stress responses. Front Plant Sci, 9: 226. |
| [24] | Sun X L, Yu Q Y, Tang L L, Ji W, Bai X, Cai H, Liu X F, Ding X D, Zhu Y M. 2013. GsSRK, a G-type lectin S-receptor-like serine/threonine protein kinase, is a positive regulator of plant tolerance to salt stress. J Plant Physiol, 170(5): 505-515. |
| [25] | Takai R, Matsuda N, Nakano A, Hasegawa K, Akimoto C, Shibuya N, Minami E. 2002. EL5, a rice N-acetylchitooligosaccharide elicitor-responsive RING-H2 finger protein, is a ubiquitin ligase which functions in vitro in co-operation with an elicitor- responsive ubiquitin-conjugating enzyme, OsUBC5b. Plant J, 30(4): 447-455. |
| [26] | Takano M, Inagaki N, Xie X Z, Yuzurihara N, Hihara F, Ishizuka T, Yano M, Nishimura M, Miyao A, Hirochika H, Shinomura T. 2005. Distinct and cooperative functions of phytochromes A, B, and C in the control of deetiolation and flowering in rice. Plant Cell, 17(12): 3311-3325. |
| [27] | Takasaki T, Hatakeyama K, Suzuki G, Watanabe M, Isogai A, Hinata K. 2000. The S receptor kinase determines self- incompatibility in Brassica stigma. Nature, 403: 913-916. |
| [28] | Tan M P, Cheng D, Yang Y N, Zhang G Q, Qin M J, Chen J, Chen Y H, Jiang M Y. 2017. Co-expression network analysis of the transcriptomes of rice roots exposed to various cadmium stresses reveals universal cadmium-responsive genes. BMC Plant Biol, 17: 194. |
| [29] | Vannini C, Locatelli F, Bracale M, Magnani E, Marsoni M, Osnato M, Mattana M, Baldoni E, Coraggio I. 2004. Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J, 37(1): 115-127. |
| [30] | Vannini C, Campa M, Iriti M, Genga A, Faoro F, Carravieri S, Rotino G L, Rossoni M, Spinardi A, Bracale M. 2007. Evaluation of transgenic tomato plants ectopically expressing the rice Osmyb4 gene. Plant Sci, 173(2): 231-239. |
| [31] | Wang Q G, Wang Y Y, Li Z, Pan J W, Wang Y F, Li Y X, Liu W. 2018. Isolation and characteristic analysis of protein kinase promoter OsSRLp from rice. Chin J Rice Sci, 32(4): 335-341. (in Chinese with English abstract) |
| [32] | Wang W S, Mauleon R, Hu Z Q, Chebotarov D, Tai S S, Wu Z C, Li M, Zheng T Q, Fuentes R R, Zhang F, Mansueto L, Copetti D, Sanciangco M, Palis K C, Xu J L, Sun C, Fu B Y, Zhang H L, Gao Y M, Zhao X Q, Shen F, Cui X, Yu H, Li Z C, Chen M L, Detras J, Zhou Y L, Zhang X Y, Zhao Y, Kudrna D, Wang C C, Li R, Jia B, Lu J Y, He X C, Dong Z T, Xu J B, Li Y H, Wang M, Shi J X, Li J, Zhang D B, Lee S, Hu W S, Poliakov A, Dubchak I, Ulat V J, Borja F N, Mendoza J R, Ali J, Li J, Gao Q, Niu Y C, Yue Z, Naredo M E B, Talag J, Wang X Q, Li J J, Fang X D, Yin Y, Glaszmann J C, Zhang J W, Li J Y, Hamilton R S, Wing R A, Ruan J, Zhang G Y, Wei C C, Alexandrov N, McNally K L, Li Z K, Leung H. 2018. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature, 557: 43-49. |
| [33] | Wilkinson L G, Tucker M R. 2017. An optimised clearing protocol for the quantitative assessment of sub-epidermal ovule tissues within whole cereal pistils. Plant Methods, 13: 67. |
| [34] | Xing S L, Li M Y, Liu P. 2013. Evolution of S-domain receptor- like kinases in land plants and origination of S-locus receptor kinases in Brassicaceae. BMC Evol Biol, 13: 69. |
| [35] | Zhang Y, Chen C, Jin X F, Xiong A S, Peng R H, Hong Y H, Yao Q H, Chen J M. 2009. Expression of a rice DREB1 gene, OsDREB1D, enhances cold and high-salt tolerance in transgenic Arabidopsis. BMB Rep, 42(8): 486-492. |
| [36] | Zhou Y B, Liu C, Tang D Y, Yan L, Wang D, Yang Y Z, Gui J S, Zhao X Y, Li L, Tang X D, Yu F, Li J L, Liu L L, Zhu Y H, Lin J Z, Liu X M. 2018. The receptor-like cytoplasmic kinase STRK1 phosphorylates and activates CatC, thereby regulating H2O2 homeostasis and improving salt tolerance in rice. Plant Cell, 30(5): 1100-1118. |
| [37] | Zou X H, Qin Z R, Zhang C Y, Liu B, Liu J, Zhang C S, Lin C T, Li H Y, Zhao T. 2015. Over-expression of an S-domain receptor-like kinase extracellular domain improves panicle architecture and grain yield in rice. J Exp Bot, 66: 7197-7209. |
| No related articles found! |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||